Abstract
The dark meal worm Tenehrio obscurus (Fabricius) is frequently found as a pest of stored grain, but it is an important resource insect like the yellow meal worm (Tenebrio molitor), larvae of which has high-protein content. The mitochondrial genome of T. obscurus is a circular molecule of 15,771 bp (GenBank accession number MG739327), with A + T bias of 72.69%. It comprises 13 protein-coding, 22 tRNA and two rDNA genes. The protein-coding genes had typical ATN (Met) as the initiation codon and were terminated by the typical stop codon TAN, except for COX2, COX3, nadh5 and nadh4. The phylogenetic tree based on maximum-likelihood method showed that the phylogenetic position of T. obscurus is closely related to T. molitor.
The dark meal worm (Tenehrio obscurus (Fabricius)) is a beetle frequently found as a pest of stored grain (Dillon and Dillon 1961), larvae of which is an important animal feed additive like the yellow meal worm (Tenebrio molitor) because of its high-protein content (Yi et al. Citation2010; Liu and Wang Citation2014). Tenehrio obscurus is very closely related to T. molitor (Plohl and Ugarković Citation1994; Rees Citation2004). There are still some clear needs for information determined for the genetic background of this beetle in both nuclei and organelles, which has not yet been determined.
Sample of adult T. obscurus was collected from Guiyang City, Guizhou Province of China, on 21 June 2017, which has been maintained in our laboratory since March 2010 (GYU accession number 201003-010). Genomic DNA isolated from the worker was fragmented for 250 bp by Covaris M220 (Covaris, Inc., Woburn, MA), built up genomic library, and sequenced (pair-end 2 × 150 bp) using Illumina Hiseq X Ten (Illumina, San Diego, CA). We obtained approximately 10,672 Mb of raw data and 10,005 Mb of clean data. Genome was de novo assembled by the SOAPdenovo v2.04 software (http://soap.genomics.org.cn/soapdenovo.html) (Luo et al. Citation2012).
The T. obscurus mitochondrial genome forms a 15,771 bp closed loop, which is biased towards A + T nucleotides, accounting for A (43.31%), T (29.38%), C (17.19%), and G (10.13%). AT-skew and GC-skew of the major strand of mt genome were approximately 0.1917 and −0.2896, respectively, using the formula proposed by Perna and Kocher (Citation1995). The length of A + T-rich region of mt genome was 1177 bp, which located between the 12S rRNA and tRNAIle.
The complete mt genome of T. obscurus contains 13 protein-coding genes (PCGs), 22 tRNA, and two rRNA. The 13 PCGs of T. obscurus used standard ATN (Met) start codon, including four ATAs (COX1, COX2, nadh5, and nadh1), four ATTs (nadh2, ATP8, nadh3, and nadh6), five ATGs (ATP6, COX3, nadh4, nadh4L, and cytB). COX1, nadh2, ATP8, ATP6, nadh4L, nadh6 and cytB genes had the common stop codon TAA; nadh3 and nadh1 had the common stop codon TAG; while COX2, COX3, nadh5 and nadh4 terminated with incomplete stop codon T completed by the addition of 3’ A residues to the mRNA. The 22 tRNA genes identified within the T. obscurus mtgenome were interspersed throughout the coding region and ranged from 59 to 69 bp in length. The size of lrRNA and srRNA genes of the T. obscurus mtgenome were 1256 and 762 bp long, respectively.
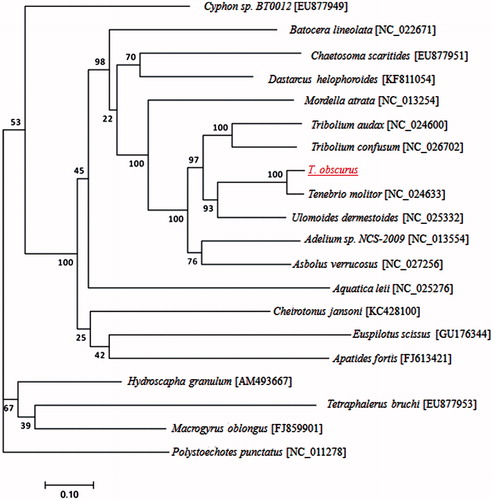
To validate the phylogenetic position of T. obscurus, 13 mitochondrial protein-coding genes of the complete mitochondrial DNA sequences from 19 closely related taxa, including 6 species from Tenebrionidae, constructed a maximum-likelihood tree using RAxML version 8.0.0 software (https://sco.h-its.org/exelixis/web/software/raxml/index.html) (Stamatakis Citation2014) (). The phylogenetic position of T. obscurus was closely clustered with T. molitor, and Ulomoides dermestoides. In conclusion, the first complete mtDNA of T. obscurus is decoded in this study and provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for Tenebrionidae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Liu LN, Wang CY. 2014. Complete mitochondrial genome of yellow meal worm (Tenebrio molitor)). Zool Res. 35:537–545.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Qi P, Liu Y. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Perna NT, Kocher TD. 1995. Unequal base frequencies and the estimation of substitution rates. Mol Biol Evol. 12:359–361.
- Plohl M, Ugarković D. 1994. Characterization of two abundant satellite DNAs from the mealworm Tenebrio obscurus. J Mol Evol. 39: 489–495.
- Rees D. 2004. Insects of stored products. CSIRO publishing.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Yi CH, He QJ, Wang L, Kuang RP. 2010. The utilization of insect-resources in Chinese rural area. J Agric Sci. 2:P146.