Abstract
Corals in the genus Porites are among the major framework builders of reef structures worldwide, yet the genus has been challenging to study due to a lack of informative molecular markers. Here, we used ezRAD sequencing to reconstruct the complete mitochondrial genome of Porites fontanesii (GenBank accession number MG754069), a widespread coral species endemic to the Red Sea and Gulf of Aden. The gene arrangement of P. fontanesii did not differ from other Scleractinia and consisted of 18,658 bp, organized in 13 protein-coding genes, 2 rRNA genes, and 2 tRNA genes. This mitochondrial genome contributes essential data to work towards a better understanding of evolutionary relationships within Porites.
Porites fontanesii Benzoni and Stefani, Citation2012 is a well-defined coral species belonging to the hard-coral family Poritiidae. Although only described recently, P. fontanesii is a common and widespread taxon in the Red Sea, with a distribution extending to the Gulf of Tadjoura, the Gulf of Aden and Socotra (Benzoni and Stefani Citation2012). The genus Porites is still taxonomically challenging in terms of species boundaries (Forsman et al. Citation2009, Citation2017; Hellberg et al. Citation2016), yet P. fontanesii is morphologically and molecularly distinctive, presenting unique morphological features, and forming a basal monophyletic clade within the Porites rDNA phylogeny (Benzoni and Stefani Citation2012).
The individual coral sample for this study was collected at Ras Qadamah reef, in Socotra Island, Yemen (12° 41.902 N; 53° 39.683 E), and is now deposited at King Abdullah University of Science and Technology, Saudi Arabia (specimen voucher SO114). Genomic DNA was extracted using DNeasy® Blood and Tissue Kit (Qiagen Inc., Hilden, Germany), quantified using Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA), and digested with frequent cutting enzymes Mbol and Sau3AI (New England Biolabs, Ipswich, MA), following Toonen et al. (Citation2013). ezRAD libraries were prepared using Illumina TruSeq® Nano DNA kit following the manufacture’s protocol, and paired-end sequenced using HiSeq® 4000 platform in the Bioscience Core Lab facility at King Abdullah University of Science and Technology, Saudi Arabia. Reads were assembled to P. lobata reference mitogenome (NC030186) using Geneious® v.10.1.3 (Biomatters Ltd. Auckland, New Zealand), and a consensus sequence exported using 0% majority option for coverage greater than 3 X. Genes were annotated using the online platforms DOGMA (Wyman et al. Citation2004) and MITOS (Bernt et al. Citation2013), and were manually inspected. tRNA was additionally scanned with the tRNAscan-SE (Schattner et al. Citation2005) web server.
The complete P. fontanesii mitogenome consisted of 18,658 bp, with the following overall base composition: A 25.81%, T 37.54%, C 13.64% and G 23.19%, in agreement with the typical mitogenome base composition (i.e. A + T rich) of scleractinian corals (Fukami and Knowlton Citation2005; Arrigoni et al. Citation2016). The reconstructed genome included 13 protein-coding genes, 2 ribosomal RNA genes (rnl and rns) and 2 transfer RNA genes (trnM and trnW). Nad5 and cox1 genes were interrupted by Group I Introns. Nad5 Group I Intron consisted of 11,135 bp, comprising 10 encoding genes, while cox1 Group I Intron was 965 bp long.
A phylogenetic tree comprising the P. fontanesii mitochondrial genome and all published mitogenomes of Poritidae and its sister taxon, Dendrophylliidae, has been reconstructed using Bayesian inference as implemented in MrBayes 3.1.2 (Ronquist and Huelsenbeck Citation2003) for 1,000,000 generations and maximum-likelihood as implemented in PhyML 3.0 (Guindon et al. Citation2003) (). The monophyly of the genus Porites is well supported by the mitochondrial phylogeny, and a distinctive position of P. fontanesii within the genus is highlighted by the reconstruction.
Figure 1. Phylogenetic reconstruction based on complete mitochondrial genomes of Porites fontanesii and other Scleractinia. Numbers at nodes represent Bayesian posterior probabilities and maximum likelihood bootstrap values. Fungiacyathus stephanus was selected as an outgroup.
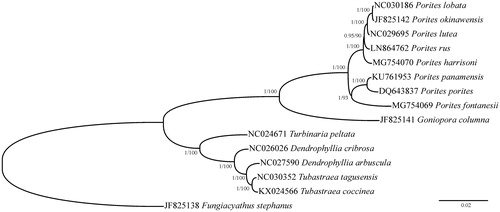
The implementation of these data with other Porites mitochondrial genomes will help clarify evolution in one of the most important framework builders of coral reefs.
Acknowledgements
F. Benzoni acknowledges Creocean, Total SA, Yemen LNG and EPA Socotra for fieldwork in Socotra.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Arrigoni R, Vacherie B, Benzoni F, Barbe V. 2016. The complete mitochondrial genome of Acanthastrea maxima (Cnidaria, Scleractinia, Lobophylliidae). Mitochondr DNA. 27:927–928. doi:10.3109/19401736.2014.926489.
- Benzoni F, Stefani F. 2012. Porites fontanesii, a new species of hard coral (Scleractinia, Poritiidae) from the southern Red Sea, the Gulf of Tadjoura, and the Gulf of Aden and its phylogenetic relationshios within the genus. Zootaxa. 3447:56–68.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Forsman Z, Knapp I, Tisthammer K. 2017. Coral hybridization or phenotypic variation? Genomic data reveal gene flow between Porites lobata and P. compressa. Mol Phylogenet Evol. 11:132–148.
- Forsman ZH, Barshis DJ, Hunter CL, Toonen RJ. 2009. Shape-shifting corals: molecular markers show morphology is evolutionarily plastic in Porites. BMC Evol Biol. 9:45.
- Fukami H, Knowlton N. 2005. Analysis of complete mitochondrial DNA sequences of three members of the Montastraea annularis coral species complex (Cnidaria, Anthozoa, Scleractinia). Coral Reefs. 24:410–417.
- Guindon S, Gascuel O, Rannala B. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by Maximum Likelihood. Syst Biol. 52:696–704.
- Hellberg ME, Prada C, Tan MH, Forsman ZH, Baums IB. 2016. Getting a grip at the edge: recolonization and introgression in eastern Pacific Porites corals. J Biogeogr. 43:2147–2159.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Toonen RJ, Puritz JB, Forsman ZH, Whitney JL, Fernandez-Silva I, Andrews KR, Bird CE. 2013. ezRAD: a simplified method for genomic genotyping in non-model organisms. PeerJ. 1:e203.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
