Abstract
The complete mitochondrial genome of T. vitrirostris was 16,975bp in length, and it contained a typically conserved structure including 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 control region (D-loop). Except ND6 and 8 tRNA genes, all other mitochondrial genes are encoded on the heavy strand, the gene arrangement was consistent with other Engraulidae mitochondrial genomes. The overall base composition is A 30.56%, C 28.40%, G 16.01%, T 25.03%. Additionally, phylogenetic tree was constructed based on 13 protein-coding genes sequences of 21 Clupeoidei species, and Chirocentrus dorab (belongs to Clupeiformes) as outgroup, the result showed that T. vitrirostris is most closely related to T. dussumieri. These results would be useful for the investigation of phylogenetic relationship, taxonomic classification, and phylogeography of the Clupeoidei.
Thryssa vitrirostris, which is belonging to Clupeoidei, Engraulidae, Thrissa, is distributed in the western Indo-Pacific. So far, the complete mitochondrial genome (mitogenome) sequence of this species has not yet been reported. In order to provide useful genetic information for genetic diversity, phylogenetic, and taxology, we determined the complete mitogenome of T. vitrirostris (GenBank accession number No. MG786488) by PCR amplification and primer walking sequence method, it would be useful for further understanding the evolution and conservation genetics of Clupeoidei.
In this study, we reported the complete mitogenome of T. vitrirostris from the South China Sea (18°3′35″N 109°24′19″ E) with accession number 20171117TV03 and identified based on the morphologic features and COI gene. Sequence alignment was conducted by BioEdit (Hall Citation1999). The complete mitochondrial genome of T. vitrirostris, with 16,975 bp in length, includes 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and 1 control region (D-Loop). The order, direction, locations of the genes and gene coding strand in the mitogenome were nearly identical to those in other Engraulidae species (Zhang and Sun Citation2013; Zhang and Xian Citation2016; Zhao and Liu Citation2016; Qu et al. Citation2017), with the exception of ND6 and eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro), all other mitochondrial genes are encoded on the H-strand. Total length of the 13 protein-coding genes was 10,902 bp, which corresponded to 64.22% of the whole mitochondrial genome. The overall base composition is A 30.56%, C 28.40%, G 16.01%, T 25.03%, AT and GC contents of mt genome are 55.58% and 44.42%, the A + T content is higher than G + C content, in common with other Clupeoidei mitogenomes (Li et al. Citation2012; Bo et al. Citation2013; Zhang et al. Citation2016). Twelve protein-coding genes start with ATG except COI with GTG. For the stop codon, nine protein-coding genes stop with TAA except ND2 with TAG, COII, ND4, and CYTB with an incomplete T––. The 22 tRNA genes vary from 66 to 75 bp in length. The T. vitrirostris mitogenome contained a small subunit rRNA (12S rRNA, located between tRNAPhe and tRNAVal genes) and a large subunit rRNA (16S rRNA, located between tRNAVal and tRNALeu genes), which were 955 and 1689 bp in length, respectively. The OL (origin of L-strand replication) is located between tRNAAsn and tRNACys, the same with other vertebrate (Bian et al. Citation2016; Guo et al. Citation2016; Zhang and Gao Citation2016), and is 20 bp in length. The control region (D-Loop) was 1327 bp in length, localized between tRNAPro and RNAPhe genes.
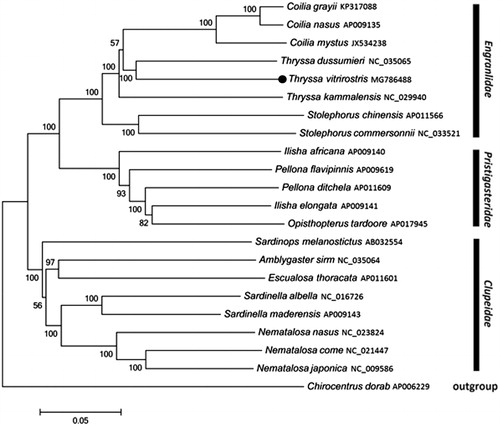
Phylogenetic tree () of T. vitrirostris and other 21 Clupeoidei species was constructed using the neighbour joining (NJ) method based on the 13 protein-coding genes. The result shows that T. vitrirostris is most closely related to T. dussumieri. We expect that the present results will further facilitate for the study on the taxonomy, population genetic structure, and phylogenetic relationships of Clupeoidei.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bian L, Su Y, Gaffney PM. 2016. The complete mitochondrial genome sequence of white perch Morone americana (Perciformes, Moronidae). Mitochondrial DNA Part B. 1:380–382.
- Bo Z, Xu T, Wang R, Jin X, Sun Y. 2013. Complete mitochondrial genome of the Osbeck’s grenadier anchovy Coilia mystus (Clupeiformes, Engraulidae). Mitochondrial DNA. 24:657–659.
- Guo M, Huang H, Gao Y. 2016. Complete mitochondrial genome of darkfin hind Cephalopholis urodeta (Perciformes, Epinephelidae). Mitochondrial DNA Part B. 1:913–916.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucl. Acids. Symp. Ser; p. 95–98.
- Li M, Zou K, Chen Z, Chen T. 2012. Mitochondrial genome of the Chinese gizzard shad Clupanodon thrissa (Clupeiformes: Clupeidae) and related phylogenetic analyses. Mitochondrial DNA. 23:438–440.
- Qu L, Ma C, Ma H, Dong Y, Wang W, Ren G, Ma L. 2017. The complete mitochondrial genome of Stolephorus commersonii. Mitochondrial DNA Part B. 2:573–574.
- Zhang J, Gao T. 2016. The complete mitochondrial genome of Thryssa kammalensis (Clupeiformes: Engraulidae). Mitochondrial DNA Part B. 1:12–13.
- Zhang N, Song N, Gao T. 2016. The complete mitochondrial genome of Coilia nasus (Clupeiformes: Engraulidae) from Ariake Sea. Mitochondrial DNA Part A. 27:1518–1519.
- Zhang BO, Sun Y. 2013. Complete mitochondrial genome of Setipinna taty (Scaly hair-fin anchovy): repetitive sequences in the control region. Mitochondrial DNA. 24:616–618.
- Zhang H, Xian W. 2016. The complete mitochondrial genome of the larvae Osbeck’s grenadier anchovy Coilia mystus (Clupeiformes, Engraulidae) from Yangtze estuary. Mitochondrial DNA Part A. 27:966–967.
- Zhao L, Liu Q. 2016. Complete mitochondrial genome sequence of Thryssa kammalensis (Clupeiformes, Engraulidae). Mitochondrial DNA Part B. 1:302–303.