Abstract
Dendrobium bellatulum Rolfe is an Endangered orchid species that is distributed in the subtropical regions of Yunnan Province, China. It was listed in the category of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES). Here, we reported the complete chloroplast (cp) genome sequence and the cp genomic features of D. bellatulum. The genome was 152,107 bp long with 129 genes comprising 83 protein-coding genes, 40 tRNA genes, and 6 rRNA genes. Phylogenetic analysis of a data set of cp genomes indicated that D. bellatulum is clustered with other species in Dendrobium.
Dendrobium bellatulum Rolfe, is an Endangered orchid species found in subtropical regions of Yunnan Province, China. It is listed in the category of the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES). The extensive use of Dendrobium materials in healthcare (Halberstein Citation2005; Shoemaker et al. Citation2005; Wojcikowski and Gobe Citation2014) resulted in severe depletion of the wild resources of Dendrobium, which has been exploited to near extinction and is now classified as one of the rare and Endangered Orchidaceae plants of China.
Chloroplast genome is much helpful in the molecular identification of Dendrobium species (Yukawa et al. Citation1996; Lau et al. Citation2001; Zhang et al. Citation2003; Asahina et al. Citation2010; Takamiya et al. Citation2011). In this study, we sequenced the complete chloroplast (cp) genome sequence and reported the cp genomic features of D. bellatulum. The specimen of D. bellatulum was collected from Yunnan Province of China and stored in Kunming Medical University Haiyuan College. The complete cp-genome sequence was submitted to GenBank under the accession number of MG595965.
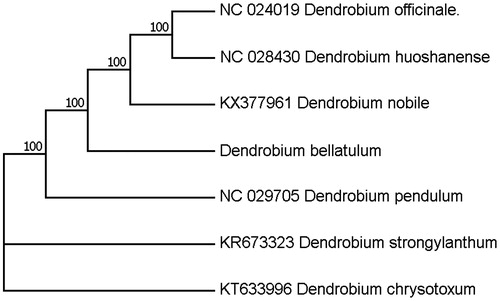
Total genomic DNA was extracted from the fresh mature leaves of D. bellatulum, and sequenced on Illumina Hiseq 2500 platform (San Diego, CA). Genome sequences were screened out and assembled with CLC genomics workbench as previously reported (Nock et al. Citation2011), which resulted in a complete circular sequence of 152,107 bp in length. The cp-genome was annotated with Dual Organellar Genome Annotator (DOGMA) (Wyman et al. Citation2004). The 152,107 bp cp-genome was made up of a large single-copy region (LSC 85,061 bp), a small single-copy region (SSC, 14,503 bp), and two inverted repeat regions (IRs, 26,297 bp). Total GC content is averagely 37.46%, of which the IR regions are 43.0% being a little bit higher than that of the LSC and the SSC regions (35.0% and 30.0%, respectively). A total of 129 genes were successfully annotated, including 89 protein-coding genes, 30 tRNA genes, and 8 rRNA genes. The tRNA genes are distributed throughout the whole genome with 17 in the LSC, 1 in the SSC, and 12 in the IR regions, while rRNAs are only situated in the IR regions. Twelve genes contained one or two introns, nine of which are protein-coding genes. To confirm the phylogenetic position of D. bellatulum, a molecular phylogenetic tree was constructed with MEGA 6.0 based on the maximum parsimony method employing a data set of the complete genome sequences of seven species from Dendrobium. The results indicated that the D. bellatulum is clustered with other species of Orchidaceae (). This newly reported chloroplast genome provides a good foundation for the molecular identification of D. bellatulum.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Asahina H, Shinozaki J, Masuda K, Morimitsu Y, Satake M. 2010. Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. J Nat Med. 64:133–138.
- Halberstein RA. 2005. Medicinal plants: historical and cross-cultural usage patterns. Ann Epidemiol. 15:686–699.
- Lau DTW, Shaw PC, Wang J, But PPH. 2001. Authentication of medicinal Dendrobium species by the internal transcribed spacer of ribosomal DNA. Planta Med. 67:456–460.
- Nock CJ, Waters DL, Edwards MA, Bowen SG, Rice N, Cordeiro GM, Henry RJ. 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol J. 9:328–333.
- Shoemaker M, Hamilton B, Dairkee SH, Cohen I, Campbell MJ. 2005. In vitro anticancer activity of twelve Chinese medicinal herbs. Phytother Res. 19:649–651.
- Takamiya T, Wongsawad P, Tajima N, Shioda N, Lu JF, Wen CL, Wu JB, Handa T, Iijima H, Kitanaka S, et al. 2011. Identification of Dendrobium species used for herbal medicines based on ribosomal DNA internal transcribed spacer sequence. Biol Pharm Bull. 34:779–782.
- Wojcikowski K, Gobe G. 2014. Animal studies on medicinal herbs: predictability, dose conversion and potential value. Phytother Res. 28:22–27.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20:3252–3255.
- Yukawa TA, Kurokawa M, Sato H, Yoshida Y, Kageyama S, Hasegawa T, Namba T, Imakita M, Hozumi T, Shiraki K. 1996. Prophylactic treatment of cytomegalovirus infection with traditional herbs. Antiviral Res. 32:63–70.
- Zhang YB, Wang J, Wang ZT, But PPH, Shaw PC. 2003. DNA microarray for identification of the herb of Dendrobium species from Chinese medicinal formulations. Planta Med. 69:1172–1174.