Abstract
The olive baboon (Papio anubis) is the most widely distributed baboon species. We report here on the complete mitochondrial genome of an olive baboon from the south-eastern edge of the species’ range from Gombe National Park (NP), Tanzania. The genome (GenBank accession number MG787545) has a length of 16,490 bp and exhibits the typical structure of mammalian mitochondrial genomes. Phylogenetically, the olive baboon from Gombe NP is most closely related to eastern P. anubis, northern P. cynocephalus and P. hamadryas. The data are an important addition to further clarify the phylogeography of baboons and phylogeny of papionins in general.
Baboons, genus Papio, are widely distributed Old World monkeys that occur over almost all sub-Saharan Africa and parts of the Arabian Peninsula. Among the six species, the olive baboon (Papio anubis) has the largest range and occurs from Mali and Guinea in the West to Eritrea in the East, and South to Uganda, Democratic Republic of Congo and Tanzania (Anandam et al. Citation2013). Phylogenetic studies on baboons using fragments of the mitochondrial genome revealed seven major clades that display a geographic pattern, but disagree with the six species taxonomy. Using these fragments, the branching pattern among the clades remained largely unresolved, while complete mitochondrial genome data revealed a well-supported phylogeny (Zinner et al. Citation2013).
We report here on the sequencing of a mitochondrial genome of an olive baboon from its southernmost range from Gombe National Park (NP), Tanzania. The skin sample (individual ID 19GNM2220916; S04°40′41″, E29°37′15″) was collected for the purpose of screening for Treponema pallidum infection in non-human primates and not specifically for this study. Collection was in accordance with Tanzanian and German laws and guidelines. DNA was extracted with methods outlined in Knauf et al. (Citation2016) and the complete mitochondrial genome was PCR amplified, sequenced and assembled following methods described in Zinner et al. (Citation2013).
The newly generated mitochondrial genome exhibits and A + T content of 56.32% and contains 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs and the control region in the order typically found in mammals (Anderson et al. Citation1981).
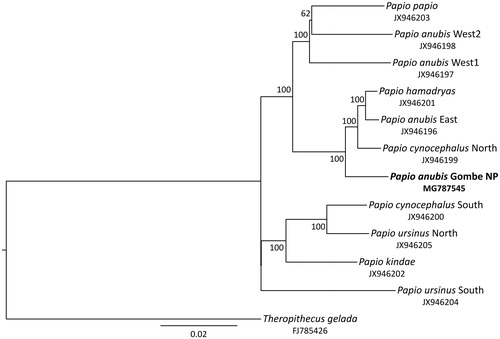
We performed phylogenetic reconstructions by adding mitochondrial genome data of other baboons representing all species and major clades (Zinner et al. Citation2009, Citation2013) and Theropithecus gelada as an outgroup. Sequences were aligned with Muscle 3.8.31 (Edgar Citation2010) in SeaView 4.5.4 (Gouy et al. Citation2010), and indels and poorly aligned positions were removed with Gblocks 0.91b (Castresana Citation2000). A maximum-likelihood tree was generated in IQ-TREE 1.5.2 (Nguyen et al. Citation2015) using the optimal substitution model (TrN + I+G) as selected by ModelFinder (Chernomor et al. Citation2016; Kalyaanamoorthy et al. Citation2017) and 10,000 ultrafast bootstrap replicates (Minh et al. Citation2013). In the obtained tree (), the olive baboon from Gombe NP forms a strongly supported (100% bootstrap) sister lineage to a clade consisting of eastern P. anubis, northern P. cynocephalus and P. hamadryas, thus, further supporting the previously detected polyphyly of olive baboons (Zinner et al. Citation2009, Citation2013).
Figure 1. Maximum-likelihood tree showing phylogenetic relationships among baboon lineages. The olive baboon from Gombe NP (highlighted in bold) clusters with eastern P. anubis, northern P. cynocephalus and P. hamadryas. Numbers on nodes refer to bootstrap values and the bar indicates substitutions per site. GenBank accession numbers are listed below species.
The mitochondrial genome of an olive baboon from Gombe NP, Tanzania is an important addition to understand the phylogeography of baboons and to further investigate the phylogeny papionins in general.
Acknowledgements
We thank the Government of the United Republic of Tanzania and its relevant institutions (Tanzania Wildlife Research Institute, Tanzania National Parks, Commission for Science and Technology) for permission and support in undertaking the study. Our gratitude goes to all Gombe National Park staff for support of the sampling activities. In the laboratory, we further thank Simone Lueert and Christiane Schwarz for their help to generate the sequence data.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Anandam MV, Bennett EL, Davenport TRB, Davies NJ, Detwiler KM, Engelhardt A, Eudey AA, Gadsby EL, Groves CP, Healy A, et al. 2013. Family Cercopithecidae (Old World monkeys) – species accounts of Cercopithecidae. In: Mittermeier RA, Rylands AB, Wilson DE, editors. Handbook of the mammals of the world, volume 3: primates. Barcelona: Lynx Edicions; p. 628–753.
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65:997–1008.
- Edgar RC. 2010. Quality measures for protein alignment benchmarks. Nucleic Acids Res. 38:2145–2153.
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27:221–224.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14:587–589.
- Knauf S, Raphael J, Mitjà O, Lejora IAV, Chuma IS, Batamuzi EK, Keyyu JD, Fyumagwa R, Lüert S, Godornes C, et al. 2016. Isolation of Treponema DNA from necrophagous flies in a natural ecosystem. EBioMedicine. 11:85–90.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30:1188–1195.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Zinner D, Groeneveld LF, Keller C, Roos C. 2009. Mitochondrial phylogeography of baboons (Papio spp.): indication for introgressive hybridization? BMC Evol Biol. 9:83.
- Zinner D, Wertheimer J, Liedigk R, Groeneveld LF, Roos C. 2013. Baboon phylogeny as inferred from complete mitochondrial genomes. Am J Phys Anthropol. 150:133–140.
