Abstract
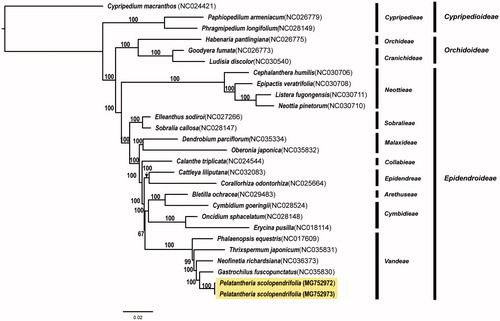
We determined the complete chloroplast genome sequence of two individuals of Pelatantheria scolopendrifolia, an endangered orchid species in Korea. The total chloroplast genome size of Mokpo (MG752972) and Naju (MG752973) population was 146,971 bp and 146,848 bp, respectively. The chloroplast genome contained 106 genes, including 72 protein-coding genes, 30 tRNA genes, and four rRNA genes. Variation in gene contents and structures was not found between two individuals. We found truncated or deleted ndh genes in P. scolopendrifolia. Phylogenetic analysis, based on the whole chloroplast genome sequences of 25 species of Orchidaceae, showed that P. scolopendrifolia was most closely related to Gastrochilus fuscopunctatus.
Pelatantheria scolopendrifolia (Makino) Aver. is a creeping evergreen epiphytic or lithophytic orchid and has small flowers (ca. 6–7 mm diameter in full blooming), leathery leaves (6–10 mm long), and many noded and branched slender stems. It occurs in East Asia, i.e. Korea, China (Tsi et al. Citation1999), and Japan (Ohasi et al. Citation2015), and with the exception of one inland population (Naju population), P. scolopendrifolia is highly restricted to southwestern coastal areas and small islands of Jellanamdo province and Jejudo Island in Korea (Lee Citation2011). Less than 10 populations are known with yearly variable population sizes. The extent of occurrence of this species was estimated to be 2000 km2 (National Institute of Biological Resources Citation2014). Given illegal collection for ornamental use and trading, habitat disturbance, and very small population sizes, P. scolopendrifolia is recognized as EN B2b(iii,iv)c(iii,iv,v) in the Korean Red List (National Institute of Biological Resources Citation2014). Despite their value and importance for conservation, very little is known for breeding system, genetic diversity, and population genetic structure of 10 known populations in Korea. In this study, we sequenced the complete chloroplast genome of two wild individuals collected from two geographical regions with different habitats, one inland population (Naju; voucher specimen CNU173926) and the other coastal population (Mokpo; voucher specimen CNU173074), in Korea. Total DNA was isolated from fresh leaves using GeneAll Exgene Plant SV mini kit (Geneall Biotechnology Co., Seoul, Korea) according to the instructions of the manufacturer. The genomic sequencing by an Illumina Miseq (Illumina Inc., San Diego, CA) platform was performed and short reads were assembled using SPAdes 3.6.1 (Bankevich et al. Citation2012) and Velvet (Zerbino and Birney Citation2008). Annotations were performed using BLAST of the National Center for Biotechnology Information and tRNAscan-SE with default setting (Lowe and Chan Citation2016). The complete chloroplast genome sequences of P. scolopendrifolia were submitted to the GenBank, and their accession numbers were acquired; coastal Mokpo population (MG752972) and inland Naju population (MG752973).
The complete chloroplast genome of coastal (MG752972) and inland population (MG752973) was 146,971 bp (LSC, 86,096 bp; SSC, 11,735 bp; IRa and IRb, 24,570 bp) and 146,848 bp (LSC, 85,984 bp; SSC, 11,730 bp; IRa and IRb, 24,567 bp), respectively. The chloroplast genome contained 106 genes, including 72 protein-coding genes, 30 tRNA genes, and four rRNA genes. We found mutations and/or deletions in 11 protein-coding genes (atpA, rpoC1, rbcL, rps18, clpP, rpoA, infA, rpl16, ndhE, ycf1, and ycf2) and two tRNA genes (trnK-UUU and trnL-UAA) between two individuals. Of 11 ndh genes in plant chloroplast genomes, four (ndhB, C, E, and J) were truncated, while the remaining seven genes were lost (most likely transferred to the mitochondrial genome; Lin et al. Citation2015). For the phylogenetic analysis, 25 complete chloroplast genome sequences of Orchidaceae were obtained and aligned using MAFFT v7.222 (Katoh and Standley Citation2013). Maximum likelihood (ML) analysis was performed using IQ-TREE v.1.4.2 (Nguyen et al. Citation2015) with 1000 bootstrap replications based on GTR +4R model. The phylogenetic tree showed that P. scolopendrifolia was closely related to Gastrochilus fuscopunctatus ().
Acknowledgements
The authors thank Dr. C. J. Oh for providing natural habitat information.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Lee NS. 2011. Illustrated flora of Korean orchid. Seoul (South Korea): Ewha Womans University Press.
- Lin CS, Chen JJ, Huang YT, Chan MT, Daniell H, Chang WJ, Hsu CT, Liao DC, Wu FH, Lin SY, et al. 2015. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci Rep. 5:9040.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- National Institute of Biological Resources. 2014. Korean red list of threatened species. 2nd ed. Incheon (South Korea): Jisungsa Publisher.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Ohasi H, Kadota Y, Murata J, Yonekura K. 2015. Wild flowers of Japan: Orchidaceae. Tokyo (Japan): Heibonsha Ltd.
- Tsi Z, Chen S, Luo Y, Zhu G. 1999. Flora Republicae Popularis Sinicae, tomus 19: Orchidaceae (3). Peking (China): Science Press.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.