Abstract
Suaeda malacosperma has limited distribution in the coastal regions of Korea and Japan and is named as a vulnerable halophyte in the Red List of Japan. The complete plastid genome of S. malacosperma is 151,989 bp long, and is composed of large single-copy (83,492 bp) and small single-copy (18,121 bp) regions plus two inverted repeats (25,188 bp each). The plastid genome encodes 130 genes, including 8 rRNAs, 37 tRNAs, and 83 protein-coding genes. rpl23 is pseudogenized. Phylogenetic analysis showed a sister relationship between Suaeda and Bienertia. This complete plastid genome is the first reported in genus Suaeda.
Suaeda malacosperma Hara is annual halophyte for which distribution is restricted to salt marshes in Korea and Japan (Shim et al. Citation2001). In Korea, it was recently reported as an unrecorded species, occurring only in brackish tidal marshes that can easily be disturbed (Shim et al. Citation2001). This species has been assigned Endangered Rank II (VU) in the Red List of Japan (https://ikilog.biodic.go.jp/Rdb/booklist). Wide swaths of habitat for halophytes in tidal regions are disappearing due to land reclamation and other human activities (Suzuki Citation2003; Millennium Ecosystem Assessment Citation2005; Choi Citation2014). Several local populations in Japan are now considered extinct (Nakanishi Citation2001). The complete plastid genome of S. malacosperma is the first report for any member of the genus Suaeda, and these details provide fundamental genetic information that could be used in developing genetic markers for conservation and in devising a phylogenomics approach for phylogenetic studies.
Total genomic DNA was extracted from silica-gel dried leaves of a single individual collected at Boseong, Korea (34°50′N, 127°24′E). The voucher specimen (J.S. Park 1610231) was deposited at the herbarium of Korea National Arboretum (KH). Genomic DNA was sequenced using the Illumina Miseq platform (LAS, Seoul, Korea). To assemble the plastid genome sequence, we generally followed the procedure of Wang and Messing (Citation2011), but with minor modifications (Choi and Choi Citation2017). The plastid genome of Bienertia sinuspersici Akhani (GenBank: KU726550) (Kim et al. Citation2016) served as our reference genome. This draft genome was annotated using DOGMA (Wyman et al. Citation2004), tRNAscan-SE (Lowe and Chan Citation2016), and the plastid genomes of B. sinuspersici, Haloxylon ammodendron (C.A. Mey.) Bunge (GenBank: KF534478), and H. persicum Bunge ex Boiss. & Buhse (GenBank: KF534479).
The plastid genome of S. malacosperma (GenBank: MG813535) is 151,989 bp long and comprises a large single-copy region (LSC; 83,492 bp), a small single-copy region (SSC; 18,121 bp), and a pair of inverted repeats (IRs; 25,188 bp) that are separated by the LSC and SSC. This genome encodes 130 genes, including 8 rRNAs, 37 tRNAs, and 83 protein-coding genes. Among these, 16 genes are duplicated in the IR region. While 17 genes contain a single intron, four others have two introns each. One gene, rpl23, is pseudogenized and known in Haloxylon (Dong et al. Citation2016). The overall GC content for S. malacosperma is 36.4%.
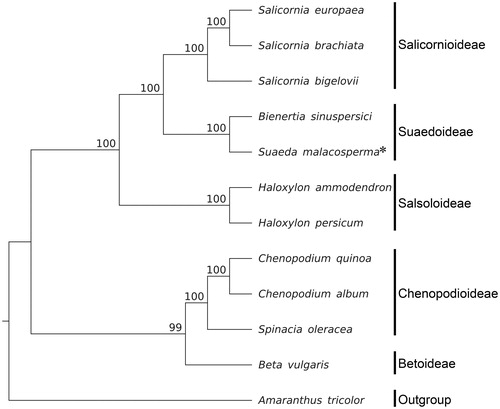
To construct a maximum likelihood (ML) phylogenetic tree, we extracted 69 genes from the genome sequences for each of 12 species within Chenopodiaceae and included Amaranthus tricolor L. of Amaranthaceae as the outgroup. The sequences were aligned using MAFFT v.7.309 (Katoh and Standley Citation2013) and the tree was analyzed with RAxML 8.2.11 (Stamatakis Citation2014) (). The topology of this ML tree for 69 genes from the plastid genomes of Chenopodiaceae members largely corresponded with that produced from a previous study based on rbcL (Kadereit et al. Citation2003). S. malacosperma was sister to B. sinuspersici.
Figure 1. Molecular phylogeny of Chenopodiaceae using 69 genes from plastid genomes of 12 species including 1 species from Amaranthaceae as outgroup. Bootstrap values are based on 1000 replicates; values are shown near each node. Plastid genome accession number is used in this phylogeny analysis: Salicornia europaea, KJ629116; S. brachiata, KJ629115; S. bigelovii, KJ629117; B. sinuspersici, KU726550; H. ammodendron, KF534478; H. persicum, KF534479; Chenopodium quinoa, KY419706; C. album, KY419707; Spinacia oleracea, AJ400848; Beta vulgaris, KR230391; Amaranthus tricolor, KX094399.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Choi IS, Choi BH. 2017. The distinct plastid genome structure of Maackia fauriei (Fabaceae: Papilionoideae) and its systematic implications for genistoids and tribe Sophoreae. PLoS One. 12:e0173766.
- Choi YR. 2014. Modernization, development and underdevelopment: reclamation of Korean tidal flats, 1950s–2000s. Ocean Coast Manage. 102:426–436.
- Dong W, Xu C, Li D, Jin X, Li R, Lu Q, Suo Z. 2016. Comparative analysis of the complete chloroplast genome sequences in psammophytic Haloxylon species (Amaranthaceae). PeerJ. 4:e2699.
- Kadereit G, Borsch T, Weising K, Freitag H. 2003. Phylogeny of amaranthaceae and chenopodiaceae and the evolution of C-4 photosynthesis. Int J Plant Sci. 164:959–986.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim B, Kim J, Park H, Park J. 2016. The complete chloroplast genome sequence of Bienertia sinuspersici. Mitochondrial DNA B Resour. 1:388–389.
- Lowe TM, Chan PP 2016. tRNAscan-SE On-line: integrating search and context for analysis of tansfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Millennium Ecosystem Assessment. 2005. Ecosystems and human well-being: synthesis. Washington (DC): Island Press.
- Nakanishi H. 2001. Phytosociological study of a Suaeda malacosperma community and distribution of Suaeda species in western Kyushu. Vegetation Science. 18:99–106. (In Japanese).
- Shim HB, Chung JY, Choi BH. 2001. One unrecorded species from Korea: Suaeda malacosperma Hara. Korean J Pl Taxon. 31:383–387. (In Korean).
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Suzuki T. 2003. Economic and geographic backgrounds of land reclamation in Japanese ports. Mar Pollut Bull. 47:226–229.
- Wang W, Messing J. 2011. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLoS One. 6:e24670.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
