Abstract
Here, we reported the complete mitochondrial genome of Cyclocheilichthys enoplos, which is widely distributed in Southeast Asia. The genome was 16,579 bp in length, containing 2 ribosomal RNA genes, 22 tRNA genes, a non-coding control region (D-loop), and 13 protein-coding genes. The overall base composition of C. enoplos was 33.1% for A, 24.5% for T, 14.9% for G, and 27.5% for C. Phylogenetic analysis showed that all Cyclocheilichthys species clustered together formed a monophyletic group. The complete mitogenome data of C. enoplos would provide a basis for further systematic studies of the genus Cyclocheilichthys.
The freshwater and potamodromous fish Cyclocheilichthys enoplos (Bleeker 1849) is a cyprinid species, which is widely distributed in Cambodia, Indonesia, Laos, Malaysia, Thailand, and Viet Nam (Froese and Pauly Citation2018). Currently, the genus Cyclocheilichthys consists of eight valid species (Froese and Pauly Citation2018), but the complete mitochondrial genome data were only available for two species.
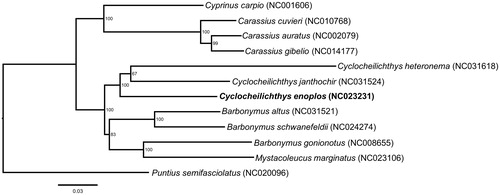
Here, we determined the complete mitogenome sequence of C. enoplos. Samples were collected from Tonle Sap Lake (13.24N/103.83E) and preserved in the museum of the Key Laboratory of Freshwater Fish Reproduction and Development (Southwest University, China). The methods for assembling the sequence followed the procedures outlined in Wang et al. (Citation2011). In order to explore the phylogenetic position of C. enoplos, we selected another 11 species for phylogenetic analysis using Puntius semifasciolatus as an outgroup (Yang et al. Citation2012). Sequences were aligned using ClustalW algorithm in MEGA7 (Kumar et al. Citation2016). RAxML version 8.2.9 (Stamatakis Citation2014) was used to construct Maximum-likelihood (ML) trees with the GTR + Γ + I model. Branch support for the resulting phylogeny was evaluated with 500 rapid bootstrapping replicates also implemented in RAxML.
The whole mitogenome of C. enoplos was a circular molecule with 16,579 nucleotides (GenBank accession number NC_023231), containing 2 ribosomal RNA genes, 22 tRNA genes, a non-coding control region (D-loop), and 13 protein-coding genes. Most of which were encoded on the heavy strand, except for ND6 and eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro). The contents of A, T, G, and C are 33.1%, 24.5%, 14.9%, and 27.5%, respectively. A + T content (57.6%) was higher than G + C content (42.4%). All of the 13 protein-coding genes show the regular initiation codon ATG with the sole exception of COI (started with GTG). Furthermore, the stop codons for 5 of the 13 protein-coding genes are TAA (ND1, ATP6, ND4L, ND5, and cyt b) and two of them are TAG (ATP8 and ND6), whereas other five genes possess incomplete stop codons TA– or T–– (ND2, ND3, ND4, COII, and COIII). Specially, COI is end with AGG.
The results from the phylogenetic analysis strongly support that the three Cyclocheilichthys species forms a monophyletic group, and C. janthochir has a close relationship with C. heteronema. However, the genus Barbonymus is not a monophyletic group because the species B. gonionotus is clustered with Mystacoleucus marginatus, rather than cluster with other Barbonymus species ().
Acknowledgements
We thank Ms Dalin Ly (Royal University of Agriculture, Cambodia) for help in field sampling.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Froese R, Pauly D, editors. 2018. FishBase. World Wide Web electronic Publication. Version (01/2018). Available from: http://www.fishbase.org
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang J, Li P, Zhang Y, Peng Z. 2011. The complete mitochondrial genome of Chinese rare minnow, Gobiocypris rarus (Teleostei: Cypriniformes). Mitochondrial DNA. 22:178–180.
- Yang L, Arunachalam M, Sado T, Levin BA, Golubtsov AS, Freyhof J, Friel JP, Chen WJ, Hirt MV, Manickam R, et al. 2012. Molecular phylogeny of the cyprinid tribe labeonini (Teleostei: Cypriniformes). Mol Phylogenet Evol. 65:362–379.