Abstract
The complete mitochondrial genome sequence plays an important role in phylogenetic studies. In this study, the complete mitochondrial genome of Monomia gladiator was obtained by Illumina and Sanger sequencing techniques. The circular genome was 15,878 bp in length, consisting of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a putative control region. This whole mitogenome composition was 33.32% for A, 35.69% for T, 11.75% for G, and 19.24% for C, respectively. The phylogenetic analysis suggested that M. gladiator was genetically closest to three Portunidae species (Charybdis japonica, C. feriata and Thalamita crenata). The newly described mitogenome may facilitate the phylogenetic studies on Portunidae crabs.
The gladiator swimming crab, Monomia gladiator was an important crab species under the family Portunidae with economic potential. It is mainly distributed in Japan, Indonesia, Thailand, India, New Zealand, Australia, and China (Zhang Citation1997). The genus Monomia has been considered as a complex species with confused nomenclature and taxonomy for a long time (Koch et al. Citation2017). Recently, molecular studies based on 16S mitochondrial gene and histone H3 fragment indicated that Portunoidea includes morphologically diverse taxa (Schubart and Reuschel Citation2009). Complete mitochondrial genome will facilitate understanding the phylogeny and species evolution in depth (Yang et al. Citation2017). However, there are only COI (Spiridonov et al. Citation2014) and partial 16S rRNA sequences (Koch et al. Citation2017) of M. gladiator available in the Genbank database. In this study, we firstly reported the complete mitochondrial genome of M. gladiator and analyzed its phylogenetic position.
Specimens of M. gladiator were collected from fish market in Xiamen City (24.463015°N, 118.087908°E), China and stored in Marine Biology Institute of Shantou University. Total genomic DNA was extracted from the muscle tissue using the described method (Ma et al. Citation2009). The complete mitogenome sequence was obtained by Illumina and Sanger sequencing methods.
The complete mitochondrial genome of M. gladiator was 15,878 bp in length (GenBank accession number: MG770549), and consisted of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes, and a putative control region. The nucleotide composition was 33.32% for A, 35.69% for T, 11.75% for G, and 19.24% for C, respectively. Of the 37 genes, 23 were encoded by the heavy strand, while 14 were encoded by the light strand. The arrangement and composition of the mitochondrial genome were similar to those of Charybdis feriata (Ma et al. Citation2015) and Thalamita crenata (Tan et al. Citation2016). The length of the 13 protein-coding genes ranged from 162 bp (ATP8) to 1728 bp (ND5). The large ribosomal RNA (16S) and small ribosomal RNA (12S) were 1352 bp and 874 bp in length, respectively. The tRNAVal was sandwiched between these two ribosomal RNA genes.
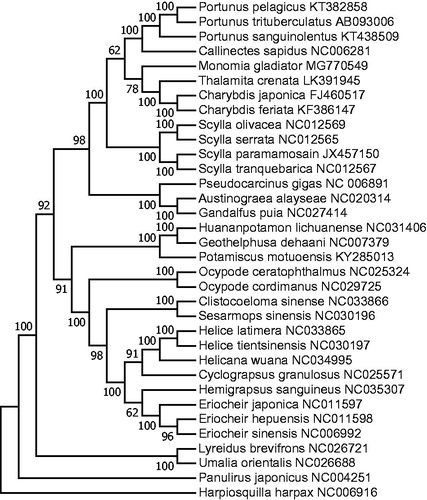
In order to determine the phylogenetic position of M. gladiator, a ML tree was constructed using the concatenated sequences of 12 protein-coding genes (except ND6) (). Hapiosquilla harpax was used as an outgroup for tree rooting. The result showed that M. gladiator was clustered together with three Portunidae species (C. japonica, C. feriata, and T. crenata), indicating that it was genetically closest to these three species. This result was also confirmed by a previous study (Spiridonov et al. Citation2014), in which M. gladiator was suggested to have closer genetic relationship to genera Charybdis and Thalamita than Portunus based on histone H3, COI, and 28S genes. The complete mitochondrial genome of M. gladiator could provide valuable information for the phylogenetic relationship of Portunidae.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Koch M, Kamanli SA, Crimmen O, Lin CW, Clark PF, Ďuriš Z. 2017. The identity of Monomia argentata (Crustacea: Brachyura: Portunidae) resolved by X-ray, computed tomography scanning and molecular comparisons. Invertebr Syst. 31:797–811.
- Ma HY, Ma CY, Li CH, Lu JX, Zou X, Gong YY, Wang W, Chen W, Ma LB, Xia LJ. 2015. First mitochondrial genome for the red crab (Charybdis feriata) with implication of phylogenomics and population genetics. Sci Rep. 5:11524.
- Ma HY, Yang JF, Su PZ, Chen SL. 2009. Genetic analysis of gynogenetic and common populations of Verasper moseri using SSR markers. Wuhan Univ J Nat Sci. 14:267–273.
- Schubart CD, Reuschel S. 2009. A proposal for a new classification of Porlunoidea and Cancroidea (Brachyura: Heterotremata) based on two independent molecular phylogenies. In: Martin JW, Crandall KA, Felder DL, editors. Crustacean. Boca Raton (FL): CRC Press; p. 533–549.
- Spiridonov VA, Neretina TV, Schepetov D. 2014. Morphological characterization and molecular phylogeny of Portunoidea Rafinesque, 1815 (Crustacea Brachyura): implications for understanding evolution of swimming capacity and revision of the family-level classification. Zool Anz. 253:404–429.
- Tan MH, Gan HM, Lee YP, Austin CM. 2016. The complete mitogenome of the swimming crab Thalamita crenata (Rüppell, 1830) (Crustacea; Decapoda; Portunidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1275–1276.
- Yang X, Ma H, Waiho K, Fazhan H, Wang S, Wu Q, Shi X, You C, Lu J. 2017. The complete mitochondrial genome of the swimming crab Chaybdis natator (Herbst) (Decapoda: Brachyura: Portunidae) and its phylogeny. Mitochondrial DNA B. 2:530–531.
- Zhang Z. 1997. The fisheries and biological characteristics of Portunus (Amphitrite) gladiator in South Fujian-Taiwan bank fishing ground. Mar Fish. 1:17–21.