Abstract
The complete mitochondrial genome sequenced from the floral egg crab Atergatis floridus (Linnaeus, 1767) and the determination of the position of the species in the reconstructed phylogenetic tree of the infraorder Brachyura using the protein coding mitochondrial genes are presented. Results show the mitochondrial genome length of A. floridus is 16,435 bp with nucleotide distribution as 33.4% A, 20.3% C, 10.5% G and 35.8% T. The structure of the complete mitochondrial genome of the species is the same as with the previous xanthid record. The result of the phylogenetic analysis suggests that A. floridus is the closest species to other Xanthidae species in the brachyuran records. This is the first complete mitochondrial genome record from the genus Atergatis.
The floral egg crab Atergatis floridus (Linnaeus Citation1767) is a cosmopolitan xanthid crab that is highly distributed in the Indo-Pacific region (Ng and Davie Citation2007). It is known to accumulate high levels of neurotoxins in its cheliped muscles (Arakawa et al. Citation1995; Saito et al. Citation2006), even in its exoskeleton and viscera (Inoue et al. Citation1968; Asakawa et al. Citation2014). In this study, the complete mitochondrial genome of A. floridus was analyzed and molecular phylogeny of the Xanthidae family in the infraorder Brachyura was investigated based on protein coding genes of the mitochondrial genome.
The A. floridus species was collected in the shores of Brgy. Amontay, Nasipit, Agusan del Norte/Philippines (8°59′25.98″N 25°19′17.97″E) on April 2017 and immediately preserved in 97% ethanol. After extraction of whole genomic DNA from the cheliped muscle, complete mitochondrial DNA analysis and the reconstruction of phylogenetic tree methods processed by following the study described previously (Karagozlu et al. Citation2016). The specimen was deposited in the Department of Biotechnology, Sangmyung University, Korea (SM00249).
The size of the complete mitochondrial genome of A. floridus was 16,180 bp (GenBank accession number: MG792341). It consists of 13 protein coding genes (PCGs), two ribosomal RNA genes, and 22 tRNAs genes. Among them 23 genes (nine PCGs and fourteen tRNAs) encoded on the majority strand and the remaining 14 genes (four PCG, eight tRNAs and two rRNAs) were encoded in the minority strand which is typical for a brachyuran species. There were three initiation codons used for protein synthesis in the mitochondrial genome namely, ATG, ATT and ATA. ATG was the most common in the protein coding genes. It is used by eight different genes (atp8, cox1, cox2, cox3, cytb, nad2, nad4 and nad4l). On the other hand, ATA was only used by the atp6 gene. As stopping codons TAA, TAG and incomplete T(AA) were used in the mitochondrial genome. The most common stopping codon identified was TAA. TAG was used by two genes (atp8 and nad2) while T(AA) was only used by the cytb gene.
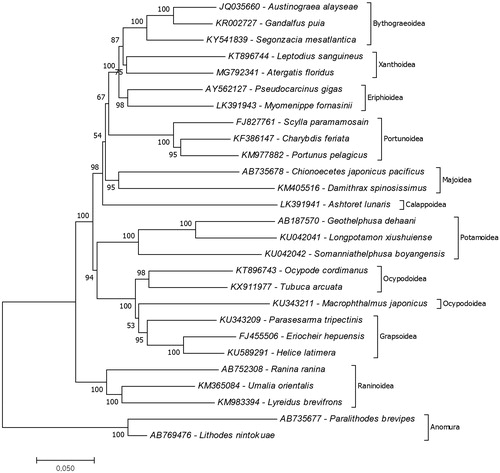
To determine the position of A. floridus in the phylogeny of brachyuran crabs, the phylogenetic tree of the different identified species were reconstructed based on the complete mitochondrial records of brachyuran species (maximum three species per superfamily) retrieved from the GenBank (). The mitochondrial genome record of Leptodius sanguineus (KT896744) was the only complete record from the xanthid crab species and was determined to be the closest to A. floridus. The results also showed the closest superfamily to Xanthoidae was Bythograeoidea. It is important to note that based from the GenBank records, the species Pseudocarcinus gigas (AY562127) belongs to the superfamily Xanthoidae. However, It was found that the species belong to the superfamily Eriphoidae thus it is suggested that there is a need for the revision in the label of this record as (WoRMS Citation2018). The present study is the first to show a complete mitochondrial genome from the genus Atergatis thus contributes to the strengthening of the brachyuran molecular genome library.
Figure 1. Molecular phylogeny of A. floridus in the infraorder Brachyura based on mitochondrial protein coding genes amino acid sequences. The data belong to A. floridus (MG792341) provided in the present study. The remained mitochondrial genome data retrieved form the GenBank. The species belongs to the infraorder Astacidea represents an outgroup for phylogenetic tree.
Disclosure statement
The authors report no conflict of interest.
Additional information
Funding
References
- Arakawa O, Nishio S, Noguchi T, Shida Y, Onoue Y. 1995. A new saxitoxin analogue from a xanthid crab Atergatis floridus. Toxicon. 33:1577–1584.
- Asakawa M, Tsuruda S, Ishimoto Y, Shimomura M, Kishimoto K, Yasuo S, Barte-Quilantang M, Gomez-Delan G. 2014. Paralytic Toxin Profiles of Xanthid Crab Atergatis Floridus Collected on Reefs of Ishigaki Island, Okinawa Prefecture, Japan and Camotes Island, Cebu Province, Philippines. Sci J Clin Med. 3:75–81.
- Inoue A, Noguchi T, Konosu S, Hashimoto Y. 1968. A new toxic crab, Atergatis floridus. Toxicon. 6:119–120.
- Karagozlu MZ, Sung JM, Lee JH, Kwon T, Kim CB. 2016. Complete mitochondrial genome sequences and phylogenetic relationship of Elysia ornata (Swainson, 1840) (Mollusca, Gastropoda, Heterobranchia, Sacoglossa). Mitochondrial DNA Part B: Resources. 1:230–232.
- Linnaeus C. 1767. Systema naturae per regna tria naturae: secundum classes, ordines, genera, species, cum characteribus, differentiis, synonymis, locis. Editio duodecima. 1. Regnum Animale. 1 & 2; p. 533–1327.
- Ng PKL, Davie PJF. 2007. On the identity of Atergatis floridus (Linnaeus, 1767) and recognition of Atergatis ocyroe (Herbst, 1901) as a valid species from the Indian Ocean (Crustacea: Brachyura: Xanthidae). Raffles Bull Zool. 16:169–175.
- Saito T, Kohama T, Ui K, Watabe S. 2006. Distribution of tetrodotoxin in the xanthid crab (Atergatis floridus) collected in the coastal waters of Kanagawa and Wakayama Prefectures. Physiol Part D: Genom Proteomics. 1:158–162.
- [WoRMS] WoRMS Editorial Board. 2018. World Register of Marine Species. VLIZ. http://www.marinespecies.org
