Abstract
Sinocyclocheilus oxycephalus is a freshwater cyprinid fish of high nutritional value, endemic to Shilin County, Southwestern China. In this study, we first sequenced the complete mitochondrial genome (mitogenome) of S. oxycephalus. The whole length of mitogenome is 16,585 bp, which contains 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and a control region. The gene arrangement and structure is identical to other previously reported Sinocyclocheilus fishes. The overall base composition is 31.26% A, 16.42% G, 25.41% T and 26.90% C, with AT content of 56.67%. Phylogenetic analysis using mitogenomes of 13 cyprinid fishes showed that S. oxycephalus are closely related to S. anophthalmus, S. grahami, and S. wumengshanensis, and 11 Sinocyclocheilus species are grouped as a monophyletic clade with strong supports.
Sinocyclocheilus oxycephalus is a freshwater cyprinid fish of high nutritional value and distributes in the Bajiang River and Heilongtan Reservoir in Shilin County, Southwestern China, which belong to mainstream Nanpanjiang River system (Li et al. Citation1985, Citation1995; Zhao and Zhang Citation2009). Here, we first report the complete mitochondrial genome (mitogenome) of S. oxycephalus, which provides the basis for future study on its systematics and conservation.
In this study, S. oxycephalus samples were collected from Heilongtan Reservoir in Shilin county (24.77°N, 103.32°E), Yunnan province, Southwestern China. The voucher specimen was preserved in 95% ethanol and deposited in the Zoological Specimen Museum of Yunnan University under accession number YNUSO20160610002). Genomic DNA from muscle tissue was extracted by DNeasy Blood & Tissue Kit (QiaGen, Valencia, CA). The DNA library was constructed and sequenced with Illumina Miseq platform (Illumina, San Diego, CA). The complete mitogenome sequence of S. oxycephalus was assembled with A5-miseq v20150522 (Coil et al. Citation2015) and SPAdes (Bankevich et al. Citation2012). The complete mitogenome was annotated using the online program DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Lowe and Eddy Citation1997).
The complete mitogenome of S. oxycephalus was deposited in the GenBank database with accession number MG686610. The whole length of mitogenome is 16,585 bp, which contains 13 protein-coding (PCGs), two ribosomal RNA (rRNA), 22 transfer RNA (tRNA) genes, and a control region (D-loop), showing 94% identities to S. grahami mitogenome (GenBank: GQ148557) (Wu et al. Citation2010). The gene arrangement and structure were identical to other Sinocyclocheilus fishes (Wu et al. Citation2010; Chen et al. Citation2017; Li et al. Citation2017). The overall base composition is 31.26% A, 16.42% G, 25.41% T and 26.90% C, with AT content of 56.67%. The ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser(UCN), tRNA-Glu, and tRNA-Pro) were encoded by the light strand, whereas the remaining genes were encoded on the heavy strand. All PCGs start with an ATG codon except for COI with start codon of GTG. Six PCGs (COI, ATP6, COIII, ND4L, ND5, and ND6) contain TAA as stop codons. Three PCGs (ND1, ND2 and ND3) end in TAG termination codon, while four PCGs (COII, ATP8, ND4 and CYTB) share incomplete stop codon T–. The 12S and 16S rRNAs are 953 bp and 1678 bp in length, respectively, located between tRNA-Phe and tRNA-Leu genes and separated by the tRNA-Val gene. Twenty-two tRNA genes can be folded into a typical cloverleaf structure and are in the range of 67–76 bp. The control region (933 bp) is located between tRNA-Pro and tRNA-Phe genes.
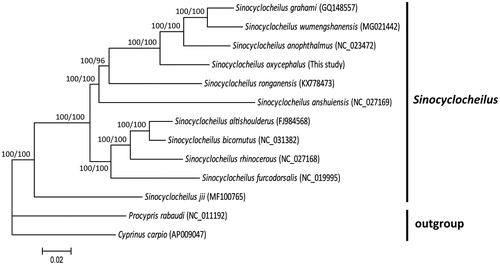
To present phylogenetic relationships among S. oxycephalus and other Sinocyclocheilus fishes, phylogenetic trees were reconstructed using MrBayes v3.2.5 (Ronquist and Huelsenbeck Citation2003) and RAxML v7.0.4 (Stamatakis Citation2006). The phylogenetic trees generated by two methods showed similar topology (). The phylogenetic results revealed that S. oxycephalus was closely related to S. anophthalmus, S. grahami, and S. wumengshanensis in Yunnan Province, and that 11 Sinocyclocheilus species were grouped as a monophyletic clade with strong supports.
Acknowledgements
The authors are grateful to Prof. Weixian Li for taxonomic identification and Mr. Tao Liu for assisting sample collection.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Chen HY, Li CQ, Chen SY, Xiao H. 2017. The complete mitochondrial genome sequence and phylogenetic position of Sinocyclocheilus wumengshanensis (Cypriniformes: Cyprinidae). Mitochondrial DNA B: Resour. 2:821–822.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31:587–589.
- Li CQ, Xiang XH, Ning T, Chen SY, Xiao H. 2017. The complete mitochondrial genome sequence of Sinocyclocheilus jii (Cypriniformes: Cyprinidae) and phylogenetic implications. Mitochondrial DNA B: Resour. 2:638–639.
- Li WX. 1985. Four new species of Sinocyclocheilus from Yunnan (Cypriniformes: Cyprinidae). Zool Res. 6:423–427.
- Li WX, Wu DF, Chen AL, Xu ZH. 1995. Analysis of nutrient composition of 2 Sinocyclocheilus in Yunnan. Chinese J Fisheries. 8:85–87.
- Lowe TM, Eddy SR. 1997. tRNA-scan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Wu XY, Wang L, Chen SY, Zan RG, Xiao H, Zhang YP. 2010. The complete mitochondrial genome of two species from Sinocyclocheilus (Cypriniformes: Cyprinidae) and a phylogenetic analysis within Cyprininae. Mol Bio Rep. 37:2163–2171.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhao YH, Zhang CG. 2009. Endemic fishes of Sinocyclocheilus (Cypriniformes: Cyprinidae) in China: Species diversity, Cave adaptation, Systematics and Zoogeography. Beijing: Science Press.