Abstract
Ancherythroculter wangi, is a unique freshwater fish and mainly distributed in the upper stream of Yangtze River and its tributary. In this study, the complete mitochondrial genome of A. wangi was first determined. The total length of the complete mitochondrial genome is 16,622 bp, contained 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, one D-loop locus, and an origin of replication on the light-strand (OL). The overall nucleotide composition was 31.19% A, 24.84% T, 27.79% C, 16.18% G, with 56.03% AT, respectively. Phylogenetic analysis both highly supported that A. wangi showed a close relationship with Culter mongolicus and A. kurematsui. These data would contribute to elucidate the evolutionary mechanisms and biogeography of Ancherythroculter and is useful for the conservation of genetics and stock evaluation for A. wangi.
Ancherythroculter wangi (Cypriniformes, Cyprinidae, Culterinae, Ancherythroculter), is an important economic freshwater fish, which mainly distributed in the upper Yangtze River and its tributaries (Ding Citation1994). Although A. wangi is popular for its delicious taste and high nutrition, unfortunately, the natural populations of this species declined rapidly in recent years due to environmental pollution, long-term overfishing, and habitat degradation (Yue et al. Citation2000).
In this study, the complete mitochondrial DNA sequence was first determined by the next generation sequencing (NGS). The specimens were collected from Neijiang, Sichuan province of China (30°16′17.05″N, 104°36′34.87″E) in September 2017, and were stored in Zoological Specimen Museum of Neijiang Normal University (accession number: 20170920BB05). A 30–40 mg fin clip was collected and preserved in 95% ethanol at 4 °C. Total genomic DNA was extracted from these caudal fins by a Tissue DNA Kit (OMEGA E.Z.N.A., Norcross, GA, USA) following the manufacturer’s protocol. Subsequently, the genomic DNA was sequenced using the NGS, and then the mitogenome was assembled using A. nigrocauda as reference.
The complete mitochondrial genome of A. wangi was a circular molecule with 16,622 bp in length (GenBank Accession numberMG 783573). The mitogenome of A. wangi contained 2 rRNA genes, 13 protein-coding genes (PCGs), 22 tRNA genes, a D-loop locus, and an origin of replication on the light-strand (OL), which was identical to the other typical vertebrates (Chen et al. Citation2016; Wang et al. Citation2017). Most of the genes were encoded on H-strand except for ND6 and 8 tRNA genes. The overall nucleotide composition was 31.19% A, 24.84% T, 27.79% C, 16.18% G, with a slight AT bias of 56.03%. All PCGs initiation codons were ATG, except for COI that began with GTG. Correspondingly, 11 PCGs stopped with the complete termination codon TAG, while the rest of PCGs ended with an incomplete termination codon T–– (COII, Cyt b), which was a little different from A. nigrocauda (ND1 with TAA) and A. lini (ND1 with TAA, ND2 and ND3 with T––, ND4, ATP6, and COIII with TA–) (Wan et al. Citation2013; Chen et al. Citation2016). Moreover, the 22 tRNA genes ranged in size from 68 bp (tRNACys) to 76 bp (tRNALeu, tRNALys). The 12S rRNA and 16S rRNA were 963 and 1691 bp, respectively. Additionally, the OL (31 bp) was located between tRNAAsn and tRNACys. Furthermore, the D-loop (938 bp) was located between tRNAPro and tRNAPhe.
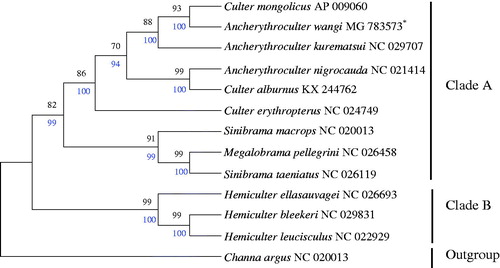
To confirm the phylogenetic relationships between A. wangi and other Culterinae subfamily fishes, phylogenetic analysis were performed on the concatenated dataset of 13 PCGs at nucleotide level with neighbour-joining (NJ) and maximum likelihood (ML) methods (Zou et al. Citation2017). The tree topologies produced by NJ had nearly a same topology as that of ML tree (). Channa argus (Channoidei, Channidae) was defined as an outgroup. The other 12 species were divided into two clades. Hemiculterella sauvagei, H. bleekeri, H. leucisculus were clustered into clade B, and the rest of species were clustered into clade A. A. wangi, Culter mongolicus, and A. kurematsui were grouped in one clade, suggested the close relationship of these species, and further confirmed that A. wangi belongs to the subfamily Culterinae. In most cases, the same genus was closer but Sinibrama taeniatus has a closer relationship with Megalobrama pellegrini than S. macrops. This situation shows that Sinibrama and Megalobrama may have descended from the same ancestor.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chen YB, Hu J, Zhao HH, Yang HR, Liu L. 2016. The complete mitochondrial genome of the Ancherythroculter lini (Cypriniformes, Cyprinidea). Mitochondrial DNA Part A. 27:3043–3044.
- Ding RH. (1994) The fishes of Sichuan, China. Chengdu (China): Sichuan Publishing House of Science and Technology; p. 203–205.
- Wan Q, Chen Y, Cheng QQ, Qiao HY. 2013. The complete mitochondrial genome sequence of Ancherythroculter nigrocauda (Cypriniformes: Cyprinidae). Mitochondrial DNA. 24:627–629.
- Wang X, Zeng S, Chen FX, Li ZY. 2017. The complete mitochondrial genome sequence of Schizothorax griseus (Cypriniformes: Cyprinidae). Mitochondrial DNA Part B. 2:648–649.
- Yue P, Shan X, Lin R. 2000. Fauna sinica: osteichthyes cypriniformes III. Beijing (China): Science Press.
- Zou YC, Xie BW, Qin CJ, Wang YM, Yuan DY, Li R, Wen ZY. 2017. The complete mitochondrial genome of a threatened loach (Sinibotia reevesae) and its phylogeny. Genes Genom. 39:767–778.