Abstract
Here, we report the complete mitochondrial genome of a Bombyx mori strain BaiyuN, which is identified to be highly resistant to Bombyx mori nucleopolyhedrovirus (BmNPV). Its complete mitochondrial genome is 15,655 bp in length (GenBank accession no. MG797555), consisting of 13 protein-coding genes, 22 tRNA genes, 2 rRNA, and 1 control region (494 bp). The complete mitogenome of the B. mori strain BaiyuN could provide a basic data for further phylogenetics and antivirus research.
The silkworm, Bombyx mori, holometabolous insect, is a key model of the Lepidoptera (Xia et al. Citation2009). BaiyuN is one strain, which was collected from Zhenjiang city, Jiangsu Province in China (N 32.20’, E 119.44’). Samples have been conserved in the national centre for silkworm genetic resources preservation, Chinese academy of agricultural sciences (SGRP, CAAS, www.cnsilkworm.com) and identified to be highly resistant to Bombyx mori nucleopolyhedrovirus (BmNPV) (Li et al. Citation2016). For better understanding of the mechanism of its antivirus, we analyzed the mitochondrial genome of B. mori BaiyuN. Total genomic DNA was extracted from the 3rd day of pupa and sequenced using the Illumina Miseq platform (Illumina lnc., San Diego, CA). A5-miseq v20150522 (Coil et al. Citation2015) and SPAdesv3.9.0 (Bankevich et al. Citation2012) software were used to assemble the obtained high quality paired-end reads. Finally, we got the complete mitochondrial genome of B. mori BaiyuN was 15,655 bp in length (GenBank accession no. MG797555), and included 13 protein-coding genes (PCGs), two rRNA genes (rrnL and rrnS), 22 tRNA genes, and a D-loop region (494 bp). The gene order and orientation of B. mori stain BaiyuN were similar to that of B. mori strains published (Zhang et al. Citation2016).
The nucleotide composition of B. mori strain BaiyuN was significantly biased (A, G, C, and T was 43.07%, 7.31%, 11.33%, and 38.29%, respectively) with A + T contents of 81.36%. The AT-skew and GC-skew of this genome were 0.059 and −0.216, respectively. Fourteen genes were transcribed on the J-strand included four PCGs (ND1, ND4, ND4L, ND5), two rRNAs, and eight tRNAs (tRNAGln, tRNACys, tRNATyr, tRNAPhe, tRNAHis, tRNAPro, tRNALeu, tRNAVal), whereas the others were oriented on the N-strand. Most of the protein-coding genes use ATN (N represents A, T, C, G) as the initiation codon whereas the cox1 gene initiated with CGA, meanwhile, all the protein-coding genes ended with TAA.
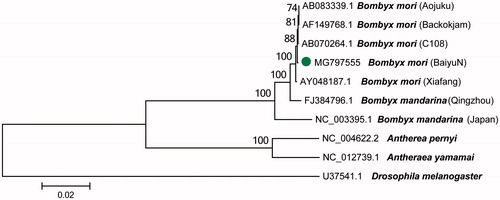
To validate the phylogenetic position of B. mori BaiyuN, we used MEGA6 software (Tamura et al. Citation2013) to construct a maximum-likelihood tree (with 500 bootstrap replicates and Kimura 2-parameter model) containing complete mitogenomes of nine species (B. mori, B. mandarina, Antheraea, respectively, and Drosophila melanogaster as a outgroup control) derived from GenBank. The mitogenome sequence of B. mori BaiyuN is similar to the sequence published by Lee et al (GenBank accession no. AF149768). So, five strains (Aojuku, Backokjam, C108, BaiyuN, Xiafang) of B. mori were first clustered together in the ML phylogenetic tree (). The mitochondrial DNA of B. mori used in different geographic varieties and different tissues suggesting that the mitochondrial DNA of B. mori are may very conservative.
Figure 1. A maximum-likelihood tree illustrating the phylogenetic position of B. mori BaiyuN among other Bombyx species. The Maximum-likelihood analysis was conducted using the complete mitogenomes, and numbers at each node are bootstrap probabilities by 500 replications shown only when they are 50% or larger. GenBank accession numbers of mitogenomic sequences for each taxon are shown.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31:587–589.
- Li G, Qian H, Luo X, Xu P, Yang J, Liu M, Xu A. 2016. Transcriptomic analysis of resistant and susceptible Bombyx mori strains following BmNPV infection provides insights into the antiviral mechanisms. Int J Genomics. 2016:2086346.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z, Dai F, Li Y, Cheng D, Li R, et al. 2009. Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science. 326:433–436.
- Zhang YL, Zhao JH, Zhou QM. 2016. The complete mitochondrial genome of Bombyx mori strain Yu39 (Lepidoptera: Bombycidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3163–3164.
