Abstract
In this study, the complete mitochondrial genome sequence of click beetle Agriotes hirayamai (GenBank accession no. MG728108) was obtained using next-generation sequencing (NGS) method. The complete mitochondrial genome of A. hirayamai is 16,156 bp in length and contains 13 protein-coding genes, 22 transfer RNAs, two ribosomal RNAs and a control region. The gene arrangement is consistent with the typical insect mitochondrial genome. Maximum likelihood tree shows that the newly sequenced A. hirayamai cluster with other two sampled species of Agriotes and the family Elateridae is monophyletic.
The family Elateridae, commonly known as click beetles due to a unique and well-known startling defence mechanism named ‘clicking’ is the largest family of Elateroidea and contains 9000 species classified to about 400 genera (Lawrence Citation1982). Here, we determined the complete mitochondrial genome of Agriotes hirayamai, which is the second mitochondrial genome sequenced to date in the genus of Agriotes.
The sampled specimen was collected from the city of Xinyang, China (the geospatial coordinates: 114.083°E, 31.833°N). The specimen was stored in the Entomological Museum of Henan Agricultural University (voucher no. MT-Zz15071207). We extracted the total genomic DNA from muscular tissue preserved in the absolute ethyl alcohol at −20 °C using the TIANamp Micro DNA Kit (Tiangen Biotech Co., Ltd., Zhongguancun, Beijing, China). We constructed the library composed of the genomic DNA of A. hirayamai and of other insects unrelated to this study using Illumina HiSeq 2500 platform (PE 150). The raw reads were de novo assembled by the SOAPdenovo software (Zhao et al. Citation2011), with an average 358.98 × coverage. We identified the complete mitochondrial genome of A. hirayamai from a single large contig (16,245 bp), by blasting the pre-determined mitochondrial cox1, cytb and rrnS gene fragments against the assembled contigs with BioEdit7.0.9.0 (Hall Citation1999).
The complete mitochondrial genome of A. hirayamai (GenBank accession no. MG728108) is 16,156 bp in length after removing the overlapping regions. The full mitochondrial genome contains 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs) and a putative control region (CR). The gene arrangement of A. hirayamai is found to be similar to most insect mitochondrial genomes (Wolstenholme Citation1992). All PCGs of A. hirayamai start with the typical start codon ATN except for nad1, which begins with the putative start codon TTG. Nine PCGs of A. hirayamai use TAA or TAG as stop codon, but four of the genes (i.e. cox2, cox3, nad4 and nad5) use T or TA as the incomplete stop codon. All 22 tRNA genes can be folded into the typical cloverleaf structure except for trnS1, in which the dihydrouracil arm cannot form a stable stem-loop structure but a simple loop. And the anticodon of trnS1 is UCU rather than usual GCU. The CR is 1490 bp in length, which consists of A + T content of 84.09% and G + C content of 15.91%.
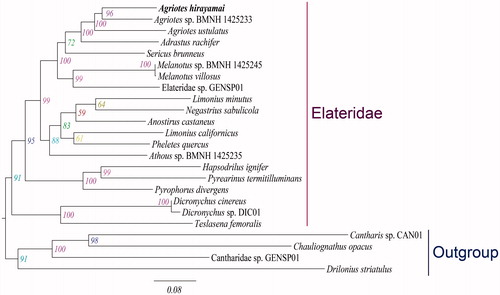
Maximum likelihood tree recovered the monophyletic Agriotes with strong bootstrap support, which was comprised of the newly sequenced A. hirayamai and other two representatives of Agriotes (). The family Elateridae was found to be monophyletic (BP = 91), which was consistent with the previous studies (Douglas Citation2011; Kundrata and Bocak Citation2011).
Figure 1. Maximum-likelihood tree inferred from the mitochondrial genome data. The maximum-likelihood analysis was reconstructed by the concatenated nucleotide sequences of 13 mitochondrial protein-coding genes (11,109 bp) using IQ-TREE (Nguyen et al. Citation2015). Numbers alongside nodes refer to bootstrap support values. The newly determined species is indicated in bold. All GenBank accession numbers for species included in this study are listed as following: Adrastus rachifer (KX087232), Agriotes hirayamai (MG728108), Agriotes sp. BMNH 1425233 (KT876879), A. ustulatus (JX412737), Anostirus castaneus (KX087237), Athous sp. BMNH 1425235 (KT876881), Cantharidae sp. GENSP01 (JX412853), Cantharis sp. CAN01 (JX412749), Chauliognathus opacus (FJ613418), Dicronychus cinereus (KX087283), Dicronychus sp. DIC01 (JX412848), Drilonius striatulus (JX412822), Elateridae sp. GENSP01 (JX412817), Hapsodrilus ignifer (KJ922149), Limonius californicus (KT852377), L. minutus (KX087306), Melanotus sp. BMNH 1425245 (KT876904), Melanotus villosus (KX087314), Negastrius sabulicola (KX087320), Pheletes quercus (KX087332), Pyrearinus termitilluminans (KJ922150), Pyrophorus divergens (EF398270), Sericus brunneus (KX087344), Teslasena femoralis (KJ938491).
Disclosure statement
All authors have read and approved the final manuscript. No conflict of interest was reported by the authors.
Additional information
Funding
References
- Douglas H. 2011. Phylogenetic relationships of Elateridae inferred from adult morphology, with special reference to the position of Cardiophorinae. Zootaxa. 2900:1–45.
- Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Kundrata R, Bocak L. 2011. The phylogeny and limits of Elateridae (Insecta, Coleoptera): is there a common tendency of click beetles to soft-bodiedness and neoteny? Zool Scr. 40:364–378.
- Lawrence JF. 1982. Coleoptera. In: Parker SP, editor. Synopsis and classification of living organisms. New York: McGraw-Hill; p. 482–553.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Wolstenholme DR. 1992. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 141:173–216.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12(Suppl 14):S2.
