Abstract
The complete chloroplast genome sequence of Sorbus ulleungensis, a recently described endemic species to Ulleung Island of Korea, was determined. The genome size was 159,632 bp in length with 36.5% GC content. It included a pair of inverted repeats (IRa and IRb) of 26,402 bp, which were separated by small single copy (SSC: 18,824 bp) and large single copy (LSC: 88,003 bp) regions. The cp genome contained 111 genes, including 78 protein coding genes, 29 tRNA genes, and four rRNA genes. Phylogenetic analysis of the combined 78 protein coding genes and four rRNA genes showed that S. ulleungensis was most closely related to Pyrus pyrifolia.
Sorbus L. sensu stricto (Rosaceae) comprises approximately 90 species and occurs widely throughout northern hemisphere (Lu and Spongberg Citation2003; McAllister Citation2005). A dense corymbs of creamy white flowers in spring followed by red, white, pink, orange or yellow foliages, and fruits in fall make this genus very attractive ornamental trees around the world. Sorbus ulleungensis Chin S. Chang, was recently described as a new endemic species on Ulleung Island, Korea (Chang and Gil Citation2014). It has been assigned originally to Sorbus commixta based on the morphological similarities, but the morphometric analysis of vegetative and reproductive traits supported taxonomic distinction between the two taxa (Chang and Gil Citation2014). This species possesses great value as garden trees as well as edible resources for Ulleung Island local people (Nitzelius Citation1989; Ong et al. Citation2016).
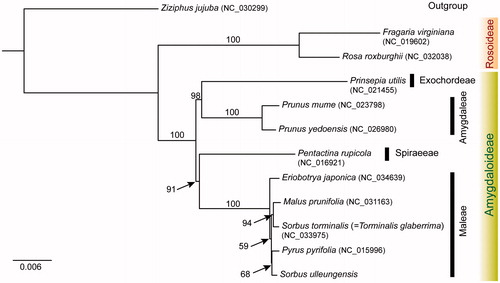
The wild individual was collected from Ulleung Island, Korea (voucher specimen SKK018558; deposited in the herbarium SKK). Total DNA was extracted from fresh leaves using the DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA). Library preparation and genomic sequencing on the Illumina Miseq platform were conducted by the Macrogen (Macrogen Inc., Seoul, South Korea). The chloroplast genome was assembled using SPAdes 2.425 and CLC Genomics Workbench v.5.5.1 (CLC Bio, Aarhus, Denmark). Then, all the contigs were aligned to the reference chloroplast genome of Sorbus torminalis (Ulaszewski et al. Citation2017) using BLAST (NCBI BLAST v2.2.31) search. The chloroplast genome of S. ulleungensis was annotated using Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004). The tRNA regions were confirmed using tRNAscan-SE with default setting (Schattner et al. Citation2005). Annotation of the start/stop codons of protein-coding genes was done using BLASTX, Geneious v.10.2.2. (Biomatters Ltd., Auckland, New Zealand), and then manually corrected for intron/exon boundaries. Maximum likelihood analysis was conducted based on concatenated 78 chloroplast coding genes using IQ-TREE v.1.4.2 (Nguyen et al. Citation2015) with 1000 bootstrap (BS) replications. The complete chloroplast genome of S. ulleungensis (GenBank accession MF695711) was 159,632 bp with 36.5% GC content and contained two inverted repeat regions (IRa and IRb) of 26,402 bp, separating a large single copy (LSC) region of 88,003 bp and a small single copy (SSC) region of 18,824 bp. The plastome contained 111 genes, including 78 protein-coding genes, 29 tRNA genes, and four rRNA genes. Eighteen genes, including seven tRNA genes and four rRNA genes, were duplicated in the IR regions. The structure, gene order, gene content, and GC content of the S. ulleungensis were similar to those of species of tribe Maleae chloroplast genomes (Bao et al. Citation2016; Ulaszewski et al. Citation2017). The phylogeny of two subfamilies (Amygdaloideae and Rosoideae) and four major tribes of Amygdaloideae of Rosaceae is shown in . Tribe Maleae of subfamily Amygdaloideae were monophyletic (100% BS, ). However, Sorbus sensu lato was not monophyletic as suggested by Lo and Donoghue (Citation2012). Sorbus ulleungensis was most closely related to Pyrus pyrifolia. A further detailed work is necessary to elucidate phylogenetic relationships among major lineages of highly complex and specious genus Sorbus.
Acknowledgements
The authors thank Dr. Joon-Mo Lee and Chung Hyun Cho for technical help during the assembly of chloroplast genome.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bao L, Li K, Liu Z, Han M, Zhang D. 2016. Characterization of the complete chloroplast genome of the Chinese crabapple Malus prunifolia (Rosales: Rosaceae: Maloideae). Conserv Genet Resour. 8:227–229.
- Chang C-S, Gil HY. 2014. Sorbus ulleungensis, a new endemic species on Ulleung Island, Korea. Harvard Pap Bot. 19:247–255.
- Lo EYY, Donoghue MJ. 2012. Expanded phylogenetic and dating analyses of the apples and their relatives (Pyreae, Rosaceae). Mol Phylogenet Evol. 63:230–243.
- Lu L, Spongberg SA. 2003. Sorbus L. In: Wu Z, Raven PH, editors. Flora of China. Vol. 9 Pittosporaceae through Connaraceae. Beijing (China): Science Press, St. Louis (MI): Botanical Garden Press; p. 144–170.
- McAllister H. 2005. The Genus Sorbus-Mountain Ash and Other Rowans. Kew (UK): Royal Botanic Gardens.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Nitzelius TG. 1989. The Ullung Rowan. Ljusterö: Föreningen för Den drologi och Parkvård.
- Ong HG, Chung JM, Jeong HR, Kim YD, Choi K, Shin CH, Lee YM. 2016. Ethnobotany of the wild edible plants gathered in Ulleung Island, South Korea. Genet Resour Crop Evol. 63:409–427.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Ulaszewski B, Sandurska E, Sztupecka E, Burczyk J. 2017. The complete chloroplast genome sequence of wild service tree Sorbus torminalis (L.) Crantz. Conserv Genet Resour. 9:419–422.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.