Abstract
It is the first report on complete chloroplast genome of Odontosoria chinensis, a traditional Chinese medicinal herb. The genome size is 156,293 bp, containing a pair of inverted repeats (IRs) (25,198 bp) separated by a small single-copy (SSC) region (20,649 bp) and a large single-copy (LSC) region (85,248 bp), respectively. The plastome has 128 genes, including 87 protein-coding genes, 32 tRNA genes, eight rRNA genes, and one pseudogene. The overall GC content is 40.43%. Maximum likelihood tree reveals that O. chinensis is basal lineages of polypods. The study will provide powerful molecular information for further phylogenetic analysis.
Odontosoria chinensis (L.) J. Sm., commonly known as Sphenomeris chinensis (L.) Maxon, belongs to Lindsaeaceae and is widespread throughout southern China at higher altitudes in evergreen forests (Dong et al. Citation2013). It is a perennially terrestrial herb with short-creeping rhizomes, pale brown stipes and rachises, and narrowly obovate ultimate segments (Dong et al. Citation2013). Except as a beautiful ornamental plant, the species is also a traditional folk medicine in China with reputation as an “all-purpose antidote” for dysentery, food and pesticide poisoning, burn injury, and incised wound (Jiangsu New Medical College Citation1977; The National Assembly Group of Chinese Herbal Medicine Citation1983). Odontosoria chinensis represents a key member to explore diversification of lindsaeoid ferns (Lehtonen et al. Citation2012) and phylogenetics and classification of Lindsaeaceae (Lehtonen et al. Citation2010; Christenhusz et al. Citation2011), acquirement of its whole chloroplast (cp) genome sequence will promote our understanding for these issues.
We collected fresh leaves of O. chinensis from South China Botanical Garden, Chinese Academy of Sciences (23°19’28.21’’N, 113°37’47.46’’E). The specimen is stored in Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 20161010). Genomic DNA was extracted using Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China) and then subjected to construct a pair-end library and sequencing in Illumina Hiseq 2500 platform (Illumina Inc., San Diego, CA). After the low-quality reads were filtered out and adaptor sequences were removed by Trimmomatic v0.32 (Bolger et al. Citation2014), approximately 1.77 G of high-quality clean data were obtained and de novo assembled by Velvet v1.2.07 (Zerbino and Birney Citation2008). The annotation was performed using the Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. Citation2004) and tRNAscan-SE programs (Lowe and Eddy Citation1997) and finally validated with BLAST searches. The circular chloroplast genome map was generated using OrganellarGenomeDraw (OGDRAW) (Lohse et al. Citation2013). Phylogenetic analysis based on maximum likelihood was performed through RAxML v8.0 (Stamatakis Citation2014) using complete cpDNA of 16 ferns including Marsilea crenata as outgroup.
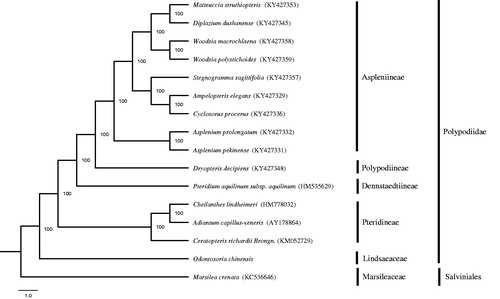
The whole chloroplast genome of O. chinensis is 156,293 bp in length with 40.43% GC content, and exhibits a typical quadripartite structure including two inverted repeats (IRa and IRb) of 25,198 bp each separated by a small single-copy (SSC) of 20,649 bp and a large single-copy (LSC) of 85,248 bp (GenBank accession number: MG913608). It possesses 128 genes including 87 protein-coding genes, 32 tRNA genes, eight rRNA genes, and one pseudogene (ndhB). Among these genes, each of IR region contains four protein-coding genes (psbA, rps7, rps12, and ycf2), five tRNA genes (trnA-UGC, trnH-GUG, trnI-GAU, trnN-GUU, and trnR-ACG), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23). In addition, 12 genes (atpF, ndhB, petB, petD, rpoC1, rpl16, rpl2, trnA-UGC, trnG-UCC, trnI-GAU, trnL-CAA, and trnV-UAC) contain one intron, while three genes (clpP, rps12, and ycf3) have two introns. GC content of the IR, SSC, and LSC is 44.17%, 36.07%, and 39.27%, respectively. ML tree reveals that O. chinensis is basal lineages of polypods (). The first complete chloroplast genome of O. chinensis will provide very powerful molecular information for further phylogenetic studies.
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Christenhusz MJM, Zhang XC, Schneider H. 2011. A linear sequence of extant families and genera of lycophytes and ferns. Phytotaxa. 19:7–54.
- Dong SY, Lin SJ, Christenhusz MJM, Barcelona J. 2013. Lindsaeaceae. In: Wu ZY, Raven PH, Hong DY, eds., Flora of China. Vol. 2–3. Beijing: Science Press; p. 139–146.
- Jiangsu New Medical College. 1977. Dictionary of Chinese Materia Medica. Vol. 1. Shanghai: Shanghai People’s Publishing House; p. 157–158. [in Chinese]
- Lehtonen S, Tuomisto H, Rouhan G, Christenhusz MJM. 2010. Phylogenetics and classification of the pantropical fern family Lindsaeaceae. Bot J Linn Soc. 163:305–359.
- Lehtonen S, Wahlberg N, Christenhusz MJM. 2012. Diversification of lindsaeoid ferns and phylogenetic uncertainty of early polypod relationships. Bot J Linn Soc. 170:489–503.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW — a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- The National Assembly Group of Chinese Herbal Medicine. 1983. Chinese Herbal Medicine. Beijing: People’s Medical Publishing House; p. 532.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.