Abstract
In this study, we sequenced the complete mitochondrial genome of Porites harrisoni using ezRAD and Illumina technology. Genome length consisted of 18,630 bp, with a base composition of 25.92% A, 13.28% T, 23.06% G, and 37.73% C. Consistent with other hard corals, P. harrisoni mitogenome was arranged in 13 protein-coding genes, 2 rRNA, and 2 tRNA genes. nad5 and cox1 contained embedded Group I Introns of 11,133 bp and 965 bp, respectively.
Keywords:
Corals of the genus Porites are among the most ecologically common corals in the tropics (Veron Citation2000). In addition to contributing to the fundamental framework for many coral reefs, corals in the genus Porites are among the most resilient to mass bleaching events, which are projected to increase under future climate conditions (Baker et al. Citation2004). Porites harrisoni (Veron Citation2000) is a major framework builder in some of the hottest seas in the world, such as the Red Sea and the Persian Arabian Gulf (Davis et al. Citation2011; Roik et al. Citation2016). Understanding the mechanisms shaping corals resistance in such extreme environments is crucial for predicting future reef scenarios (Voolstra et al. Citation2015).
Here, we sequenced the complete mitochondrial genome of P. harrisoni (GenBank accession number MG754070) using restriction site associated DNA (RAD) sequencing. The coral sample used in this study was collected at Dhi Dahaya Reef in the southern Saudi Arabian Red Sea (N 16°52.377′, E 41°26.409′), deposited at King Abdullah University of Science and Technology, Saudi Arabia (voucher number SA1704), and the coral tissue was preserved in 98% EtOH. DNA extraction was performed with DNeasy® Blood and Tissue Kit (Qiagen Inc., Hilden, Germany) and quantification was performed with Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA). We used ezRAD sequencing method, and MboI and Sau3AI (New England Biolabs, Ipswich, MA) frequent cutters. ezRAD libraries have been prepared with TruSeq® Nano DNA kit, and paired-end sequenced with the HiSeq® 4000 platform in the Bioscience Core Lab facility at King Abdullah University of Science and Technology, Saudi Arabia. The obtained reads were assembled to the deposited reference mitogenome of Porites lobata (NC030186) using Geneious® v.10.1.3 (Biomatters Ltd. Auckland, New Zeland). A 0% majority option for coverage greater than 3× was applied when calling a consensus sequence, which assembled to 95.2% of P. lobata genome. Genes were annotated with DOGMA (Wyman et al. Citation2004) and MITOS (Bernt et al. Citation2013), and manually verified.
The complete mitogenome consisted of 18,630 bp, with a base composition of 25.92% A, 13.28% T, 23.06% G, and 37.73% C. The total number of protein-coding genes was 13, consistent with the typical scleractinian mitogenome organization (Medina et al. Citation2006; Fukami et al. Citation2007). A total of two ribosomal RNA (rnl, nrs) and two transfer RNA (trnM, trnW) genes was recovered. Among the 13 protein-coding genes, nad5 and cox1 were interrupted by Group I Introns, 11,133 bp and 965 bp in length, respectively.
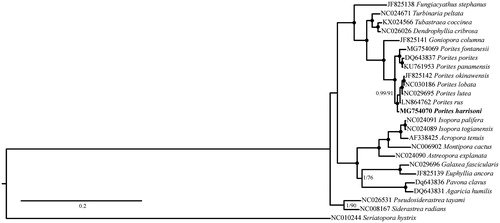
Porites harrisoni phylogenetic position with reference to other Scleractinia has been reconstructed using Bayesian inference as implemented in MrBayes 3.1.2 (Ronquist and Huelsenbeck Citation2003) for 4,000,000 generations and maximum-likelihood as implemented in PhyML 3.0 (Guindon et al. Citation2003) with 1000 bootstrap replicates. The mitogenomes phylogeny shows that all Porites sequences, including P. harrisoni cluster in a single group ().
Figure 1. Phylogenetic reconstruction based on complete mitochondrial genomes of P. harrisoni (in bold) and other Complex scleractinian corals. Numbers at nodes represent Bayesian posterior probabilities (Bp) and maximum likelihood bootstrap (ML) values. Black dots represent Bp = 1 and ML = 100. Seriatopora hystrix was selected as outgroup.
The complete mitochondrial genome of P. harrisoni will provide a baseline for further studies aimed at a better understanding of the evolution and adaptation of Scleractinia.
Acknowledgements
This research was undertaken in accordance with the policies and procedures of KAUST. Permissions relevant for KAUST to undertake the research have been obtained from the applicable governmental agencies in the Kingdom of Saudi Arabia. ZHF would like to thank the Seaver Institute for support. This is HIMB contribution number 1715 and SOEST contribution number 10310.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Baker AC, Starger CJ, McClanahan TR, Glynn PW. 2004. Coral reefs: corals' adaptive response to climate change. Nature. 430:741.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Davis KA, Lentz SJ, Pineda J, Farrar JT, Starczak VR, Churchill JH. 2011. Observations of the thermal environment on Red Sea platform reefs: a heat budget analysis. Coral Reefs. 30:25–36.
- Fukami H, Chen CA, Chiou CY, Knowlton N. 2007. Novel group I introns encoding a putative homing endonuclease in the mitochondrial cox1 gene of scleractinian corals. J Mol Evol. 64:591–600.
- Guindon S, Gascuel O, Rannala B. 2003. A simple, fast, and accurate qlgorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Medina M, Collins AG, Takaoka TL, Kuehl JV, Boore JL. 2006. Naked corals: skeleton loss in Scleractinia. Proc Natl Acad Sci USA. 103: 9096–9100.
- Roik A, Röthig T, Roder C, Ziegler M, Kremb SG, Voolstra CR. 2016. Year-long monitoring of physico-chemical and biological variables provide a comparative baseline of coral reef functioning in the Central Red Sea. Magar V, editor. PLoS One. 11:e0163939.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Veron JEN 2000. Corals of the World. Australia: Australian Institute of Marine Science and CRR Qld Pty Ltd.
- Voolstra CR, Miller DJ, Ragan MA, Hoffmann AA, Hoegh-Guldberg O, Bourne DG, Ball EE, Ying H, Foret S, Takahashi S, et al. 2015. The ReFuGe 2020 Consortium – using “omics” approaches to explore the adaptability and resilience of coral holobionts to environmental change. Front Mar Sci. 2:68.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
