Abstract
Asian palm civet (Paradoxurus hermaphroditus) is one of the smallest palm civet which is least studied. Here, we report the first complete mitochondrial (mt) genome of Asian palm civet (P. hermaphroditus). The circular mt genome with a length of 16,706 bp contained 1 control region, 2 rRNAs, 13 protein-coding genes, and 22 tRNAs. Overall base composition of the complete mt DNA was 33.7% A, 30.5% T, 22.9% C, and 12.9% G. All the genes in mt genome of Asian palm civet (P. hermaphroditus) were distributed on the H-strand, except ND6 and eight tRNA genes encoded on the L-strand. Maximum likelihood (ML) and Bayesian inference (BI) methods were used to infer the phylogenetic relationship of P. hermaphroditus. The phylogenetic analysis shows that all species from the family Viverridae cluster together, in which P. hermaphroditus exhibits the closest relationship with P. larvata.
The subfamily Viverridae comprises of five genera: Paradoxurus, Paguma, Arcticis, Arctogalidia, and Macrogalidia that are found in South and Southeast Asia (Wozencraft Citation2005). Till date, only five complete mitochondrial genomes of viverrids have been reported (Zhang et al. Citation2015; Hassanin and Veron Citation2016; Hassanin Citation2016; Weng et al. Citation2016; Salleh et al. Citation2017). Here, we report the first complete mitochondrial (mt) genome sequence of Asian palm civet (Paradoxurus hermaphroditus, GenBank Accession No. MG200264).
Primary fibroblasts culture established as described by Yelisetti et al. (Citation2016), from post-mortem tissue of a male adult Asian palm civet from Nehru Zoological Park, Hyderabad, India (N: 17°21′04″E: 78°26′59″) and maintained at LaCONES Genome Bank (LGB-PC-001), was used for DNA extraction by standard phenol-chloroform isoamyl alcohol method (Sambrook et al. Citation1989). PCR amplification and sequencing of the entire mt genome was done using three pairs of primers with an average amplicon size of 5–6 kb each.
The total length of the P. hermaphroditus mt DNA was 16,706 bp. It contained 1 control region (D-loop), 2 rRNA genes, 13 protein-coding genes, and 22 tRNA genes. Origin of reading frame of all protein-coding genes and gene orders were identical to other members of Viverridae. The overall base composition of A, T, G, and C was 33.7%, 30.5%, 12.9%, and 22.9%, respectively and the GC content of the mt genome was 35.8%. Most of the P. hermaphroditus genes were encoded on the H-strand, except for ND6 gene and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, tRNAPro). The control region was located between tRNAPro and tRNAPhe. The length of D-loop was 1321 bp ranging from 15,386–16,706 bp. Most of the protein-coding genes had ATG (Met) as start codon except ND2, ND3, and ND5 (ATT). Further, ND1, ATP8, ND4, Cyt b ended with TAG; COX1, COX2, ATP6, ND5, and ND6 ended with TGA, ND2, ND3, ND4l, and COX3 ended with TAA as stop codons.
For sequence comparison, mt genomes of five species of viverrids were obtained from NCBI GenBank. The mt genome of Asiatic lion (Panthera leo persica, Felidae, Tabasum et al. Citation2016) was used as an outgroup. The phylogenetic relationships were constructed among all viverrids based on 13 protein-coding genes. The best model of sequence evolution for each gene in both the analysis was obtained by jModeltest 2.1.5 (Darriba et al. Citation2012). Bayesian analysis (BI) was performed using Mr Bayes v 3.2 (Ronquist et al. Citation2012) with four chains of 1.1 × 105 generations and sampling the trees every 100 generations. Maximum-likelihood (ML) analysis was performed using MEGA 7 (Kumar et al. Citation2016).
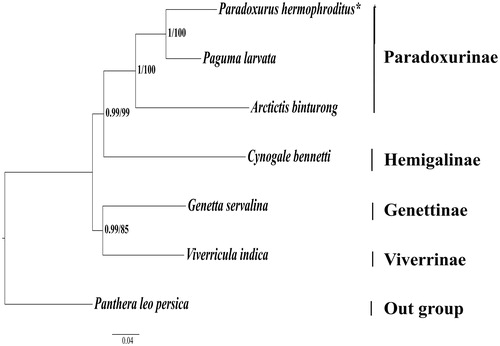
All species of Viverridae clustered together, and P. hermaphroditus exhibited the closest relationship with P. larvata () in consistence with previous reports (Patou et al. Citation2010, Veron et al. Citation2015, Citation2017). Since, there is a limited genetic data available on Asian palm civet (P. hermaphrodites) from Indian sub-continent, the present study will contribute significantly to in-depth phylogenetic studies and proper assessment of conservation status of this species.
Figure 1. Phylogenetic relationships among the Viverrids. Clade support values are given by Bayesian posterior probabilities/ML bootstrap percentages. The phylogenetic tree was rooted using P. leo persica (KU234271). The analyzed species and their respective NCBI accession number are as follows: Paguma larvata (NC_029403), Arctictis binturong (KY117560), Cynogale bennetii (KY117544), Genetta servalina (NC_024568), and Viverricula indica (NC_025296). *P. hermaphroditus (MG200264 – this study).
Nucleotide sequence accession number
The complete mitogenome sequence of Asian palm civet (Paradoxurus hermaphroditus) has been assigned Genbank accession number MG200264.
Acknowledgements
The authors would like to thank Director, CCMB and CSIR, India for all the support, Dr B Sambasiva for providing the fibroblast culture.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Darriba D, Taboada GL, Doalla R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Hassanin A. 2016. The complete mitochondrial genome of the Servaline Genet, Genetta servalina, the first representative from the family Viverridae (Mammalia, Carnivora). Mitochondrial DNA Part A. 27:906–907.
- Hassanin A, Veron G. 2016. The complete mitochondrial genome of the Spotted Linsang, Prionodonpardicolor, the first representative from the family Prionodontidae (Mammalia, Carnivora). Mitochondrial DNA Part A. 27:912–913.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Patou ML, Wilting A, Gaubert P, Esselstyn JA, Cruaud C, Jennings AP, Fickel J, Veron G. 2010. Evolutionary history of the Paradoxurus palm civets–a new model for Asian biogeography. J Biogeogr. 37:2077–2097.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Salleh FM, Ramos-Madrigal J, Peñaloza F, Liu S, Sinding MH, Patel RP, Martins R, Fickel DL, Roos C, Shamsir MS, Azman MS. 2017. An expanded mammal mitogenome dataset from Southeast Asia. GigaScience. 6:1–8.
- Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. 2nd edition. New York: Cold Spring Harbor Laboratory Press.
- Tabasum W, Ara S, Rai N, Thangaraj K, Gaur A. 2016. Complete mitochondrial genome sequence of Asiatic lion (Panthera leo persica). Mitochondrial DNA Part B. 1:619–620.
- Veron G, Bonillo C, Hassanin A, Jennings AP. 2017. Molecular systematics and biogeography of the Hemigalinae civets (Mammalia, Carnivora). Europ J Taxon. 285:1–20.
- Veron G, Patou ML, Tóth M, Goonatilake M, Jennings AP. 2015. How many species of Paradoxurus civets are there? New insights from India and Sri Lanka. J Zool SystEvol Res. 53:161–174.
- Weng HM, Wang L, Chan FT, Sun PY, Li KY, Ju YT. 2016. The complete mitochondrial genome of the small Indian civet, Viverricula indica taivana–the first complete representation of the genus Viverricula. Mitochondrial DNA Part A. 27:1650–1651.
- Wozencraft WC. 2005. Order carnivora. In Wilson DE & Reeder, DM, editors. Mammal species of the world: Third Edition. Baltimore: The Johns Hopkins University Press. p. 532–628.
- Yelisetti UM, Komjeti S, Katari VC, Sisinthy S, Brahmasani SR. 2016. Interspecies nuclear transfer using fibroblasts from leopard, tiger, and lion ear piece collected postmortem as donor cells and rabbit oocytes as recipients. In Vitro Cell Dev Biol Anim. 52:632–645.
- Zhang D, Xu L, Bu H, Wang D, Xu C, Wang R. 2015. The complete mitochondrial genome of the masked palm civet (Paguma larvata, Mammalia, Carnivora). Mitochondrial DNA, Part A. 27:3764–3765.
