Abstract
Trachidermus fasciatus is a small catadromous fish and has been listed as a second class state protected aquatic animal since 1988 in China due to the declines in its abundance. We describe the complete mitogenome of T. fasciatus in this study. The mitogenome is 16,536 nucleotides long and contains 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes, and two main non-coding regions. The overall base composition includes C (30%), A (26.3%), T (25.5%), and G (18.2%). Moreover, the 13 PCGs encode 3800 amino acids in total, all the PCGs use the initiation codon ATG except COI uses GTG. Most of them have TAA or TAG as the stop codon, except COII, ND4 and Cytb use an incomplete stop codon T. The phylogenetic tree based on the neighbour joining method was constructed to provide relationship within Cottidae, which could be a useful basis for management of this species.
The Roughskin sculpin, Trachidermus fasciatus is the only member of the monotypic genus Trachidermus. Formerly, they were very abundant along the coast of China, the pollution from rapid urban development, overfishing and the construction of dams and dikes threaten the species so they are now threatened with extinction (Bi et al. Citation2011). In this study, we determined the complete mitochondrial genome of T. fasciatus and explored the phylogenetic relationship based on 12 mitochondrial protein-coding genes (PCGs) located on heavy strand of 18 Cottidae species, contributing to additional information of conservation for this specie (Li et al. Citation2016).
Sample was collected from Fuchun River in Fuyang, China (27°56′17″N; 119°51′15″E). Specimen was stored in laboratory of Zhejiang Ocean University with accession number 20150826SJL22. Twelve primer pairs were designed to amplify the complete mitogenome. DNA sequences were assembled by Codoncode Aligner. The calculation of base composition and a neighbour joining (NJ) tree were constructed by MEGA5.0 software (Tamura et al. Citation2011).
The entire genome sequence is 16,536 bp in length (GenBank accession MG784718), consisting of 13 PCGs, 22 tRNA genes, two rRNA genes, one replication origin (OL) and a control region (CR). The overall base composition is 30% A, 26.3% C, 25.5% T, and 18.2% G. Twelve PCGs, 14 tRNA genes, and two rRNA genes were located on the heavy strand, while one PCG (ND6) and eight tRNA genes (tRNA Gln, RNA Ala, tRNA Asn, tRNA Cys, tRNA Tyr, tRNA Ser, tRNA Glu, and tRNA Pro) on the light strand. Thirteen PCGs encode 3800 amino acids in total. All the PCGs use the initiation codon ATG except COI use GTG, which is quite common in other teleost mtDNA (Sergei et al. Citation2013; Balakirev et al. Citation2016; Fast et al. Citation2017). Most of them have TAA or TAG as the stop codon, except COII, ND4 and Cytb use an incomplete stop codon T. A total of 52 base pairs in 11 intergenic spacers are found ranging from 1 to 30 bp in length. Eleven overlapping areas (32 bp in total) are observed, notable overlapping occurred at three pairs of PCGs. ATP8 and ATP6 overlapped by 10 nucleotides, ND4L and ND4 by 7 bp, ND5 and ND6 by 4 bp.
The lengths of 12S rRNA located between tRNAPhe and tRNAVal and 16S rRNA located between tRNAVal and tRNALeu were 945 bp and 1690 bp, respectively (Wang and Xiuying Citation2008). The origin of light-strand replication (OL) is located in a cluster of five tRNA genes (WANCY) and the CR, with 860 bp, is located between the tRNA-Pro and tRNA-Phe genes (Zhang et al. Citation2015).
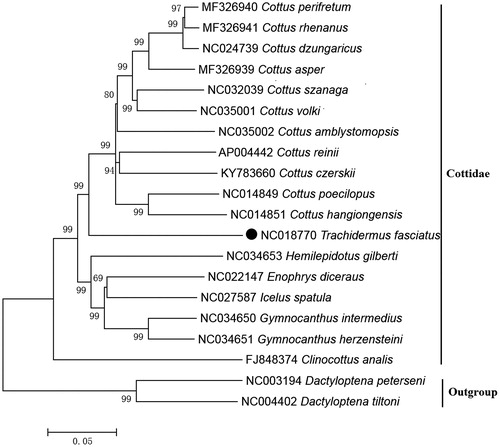
The NJ tree was constructed based on 12 mitochondrial PCGs located on heavy strand of 18 Cottidae species. The results of the present study supports T. fasciatus has a closest relationship with Cottus with a bootstrap probability of 99% (), which is in accord with the results based on other molecular methods (Yokoyama and Goto Citation2005).
Figure 1. Neighbour joining (NJ) tree of 18 Cottidae species based on 12 PCGs. The bootstrap values are based on 10,000 resamplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labelled with a black spot.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Balakirev ES, Saveliev PA, Ayala FJ. 2016. Complete mitochondrial genome of the Amur sculpin Cottus szanaga (Cottoidei: Cottidae). Mitochondrial DNA B. 1:737–738.
- Bi X, Yang Q, Gao T, Li C. 2011. The loss of genetic diversity during captive breeding of the endangered sculpin, Trachidermus fasciatus, based on ISSR markers: implications for its conservation. Chin J Oceanol Limnol. 29:958–966.
- Fast K, Aguilar A, Nolte AW, Sandel MW. 2017. Complete mitochondrial genomes for Cottus asper, Cottus perifretum, and Cottus rhenanus (Perciformes, Cottidae). Mitochondrial DNA B. 2:666–668.
- Li Y-L, Xue D-X, Gao T-X, Liu J-X. 2016. Genetic diversity and population structure of the roughskin sculpin (Trachidermus fasciatus Heckel) inferred from microsatellite analyses: implications for its conservation and management. Conserv Genet. 17:921–930.
- Sergei V, Shedko ILM, Nemkova GA. 2013. Complete mitochondrial genome of the poorly known Amur sculpin Mesocottus haitej (Cottoidei: Cottidae). Mitochondrial DNA. 26:147–148.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Wang D, Xiuying W. 2008. A study on hydrological and hydraulic features of Trachidermus fasciatus migration. Adv Water Res Hydraulic Eng. 2:562–567.
- Yokoyama R, Goto A. 2005. Evolutionary history of freshwater sculpins, genus Cottus (Teleostei; Cottidae) and related taxa, as inferred from mitochondrial DNA phylogeny. Mol Phylogenet Evol. 36:654–668.
- Zhang H, Zhang Y, Qin G, Lin Q. 2015. The complete mitochondrial genome sequence of the network pipefish (Corythoichthys flavofasciatus) and the analyses of phylogenetic relationships within the Syngnathidae species. Mar Genomics. 19:59–64.
