Abstract
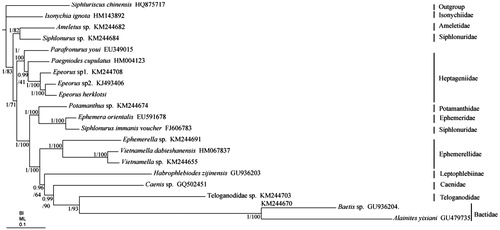
The mitochondrial genome of Epeorus herklotsi (Ephemeroptera: Heptageniidae) is a circular molecule of 15,801 bp in length with a base composition of 32.7% A, 32.9% T, 21.5% C, 13.0% G, including extra tRNAMet gene. The IMQM tRNA cluster is found in E. herklotsi as well as Parafornuru youi and two species of Epeorus (KM244708, KJ493406), while the typical IQM tRNA cluster is found in Paegniodes cupulatus. In BI and ML phylogenetic trees, the monophyly of the families Heptageniidae, Baetidae, and Ephemerellidae are highly supported. E. herklotsi is a sister clade to Epeorus sp2. (KJ493406).
The phylogenetic relationships of Ephemeroptera using morphological or molecular methods still exists disputes (Kristensen Citation1981; Ogden and Whiting Citation2003; Zhang et al. Citation2008; Simon and Hadrys Citation2013; Li et al. Citation2014; Misof et al. Citation2014). Thirteen complete mitochondrial genomes and six partial mitochondrial genomes of Ephemeroptera are available (Zhang et al. Citation2008; Li et al. Citation2014; Tang et al. Citation2014; Zhou et al. Citation2016). In the 19 known mayfly mitochondrial genomes, gene rearrangements are found in five species (e.g. Parafronurus youi, Epeorus sp1., Epeorus sp2., Siphluriscus chinensis, Alainites yixiani), especially in Heptageniidae two types of IQM and IMQM tRNA cluster were found. Hence, we sequenced the mitochondrial genome of Epeorus herklotsi (Ephemeroptera: Heptageniidae) to analyse the characteristics of mitochondrial gene arrangement and to discuss the phylogenetic relationships within Ephemeroptera.
The samples of E. herklotsi identified by Dr. JY Zhang were collected in Lishui, Zhejiang province, China. All samples of E. herklotsi were stored in Lab of HY Cheng, College of Chemistry and Life Science, Zhejiang Normal University. And the total genomic of DNA was isolated from one leg of E. herklotsi using Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China). The conserved primers and modified primers for PCR amplification were designed according to Zhang et al. (Citation2008). PCR products were sent to Sangon Biotech Company for sequencing with both strands.
The mt genome of E. herklotsi is a circular molecule of 15,801 bp in length, with the overall base composition of 32.7% A, 32.9% T, 21.5% C, 13.0% G, including extra tRNAMet gene. The gene rearrangement with IMQM tRNA cluster was found in E. herklotsi, which is similar to P. youi (Zhang et al. Citation2008) and two species of Epeorus (KM244708, KJ493406) (Tang et al. Citation2014), while is different to Paegniodes cupulatus with the typical IQM tRNA cluster (Zhou et al. Citation2016).
Bayesian inference (BI) and maximum likelihood (ML) trees were constructed using the 13 PCGs from 20 species using S. chinensis (Li et al. Citation2014) as outgroup (). Each alignment was performed by Gblock 0.91b (Castresana Citation2000) using default settings in order to select conserved regions of the nucleotide. BI and ML analysis were performed by MrBayes3.1.2 (Huelsenbeck and Ronquist Citation2001) and RAxML 8.2.0 (Stamatakis Citation2014), respectively. In BI and ML phylogenetic trees, the monophyly of the families Heptageniidae, Baetidae, and Ephemerellidae is highly supported (), but the monophyly of Siphlonuridae is failed. Siphlonurus sp. (Siphlonuridae) is a clade sister to Ameletus sp. (Ameletidae), however, Siphlonurus immanis (Siphlonuridae) is a sister clade to Ephemera orientalis (Ephemeridae). The monophyly of Epeorus is highly supported and E. herklotsi is a sister clade to Epeorus sp2. (KJ493406). In addition, Baetidae (Baetis sp. + Alainites yixiani) is a sister clade to Teloganodidae which is not consist with view of Ogden and Whiting (Citation2005). We found the long branch attraction existed in Baetidae, which may affect the phylogenetic relationship of Baetidae.
Figure 1. Phylogenetic tree of the relationships among 20 species of Ephemeroptera, including E. herklotsi based on the nucleotide dataset of the 13 mitochondrial protein-coding genes. The Bayesian posterior probability values and the maximum-likelihood bootstrap values are indicated above nodes. The GenBank numbers of all species are shown in the figure.
Acknowledgements
The authors would like to thank Ya-Jie Gao for aid in taxon sampling.
Nucleotide sequence accession number: The complete mitochondrial genome of E. herklotsi has been assigned the GenBank accession number MG870104.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Kristensen NP. 1981. Phylogeny of insect orders. Annu Rev Entomol. 26:135–157.
- Li D, Qin JC, Zhou CF. 2014. The phylogeny of Ephemeroptera in Pterygota revealed by the mitochondrial genome of Siphluriscus chinensis (Hexapoda: Insecta). Gene. 545:132–140.
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science. 346:763–767.
- Ogden TH, Whiting MF. 2003. The problem with “the Paleoptera problem” sense and sensitivity. Cladistics. 19:432–442.
- Ogden TH, Whiting MF. 2005. Phylogeny of Ephemeroptera (mayflies) based on molecular evidence. Mol Phylogenet Evol. 37:625–643.
- Simon S, Hadrys H. 2013. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol Phylogenet Evol. 69:393–403.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tang M, Tan M, Meng G, Yang S, Su X. 2014. Multiplex sequencing of pooled mitochondrial genomes a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166.
- Zhang J, Zhou C, Gai Y, Song D, Zhou K. 2008. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene. 424:18–24.
- Zhou D, Wang YY, Sun JZ, Han YK. 2016. The complete mitochondrial genome of Paegniodes cupulatus (Ephemeroptera: Heptageniidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:925–926.
