Abstract
In this study, the complete mitogenome sequence of Montipora digitata (Cnidarian: Acroporidae) has been decoded for the first time using the low-coverage whole-genome sequencing method. The overall base composition of M. digitata mitogenome for A, C, G, and T is 24.9%, 14.2%, 24.2%, and 36.8%, respectively, with a GC content of 38.3%. The assembled mitogenome consists of 17,884 bp with unique 13 unique protein-coding genes (PCGs), two ribosomal RNAs, and only three transfer RNAs genes. M. digitata has one big intron insertion in the ND5 gene. The complete M. digitata mitogenome provides essential and important molecular DNA data for further phylogenetic and evolutionary stony coral analysis.
Montipora digitata belongs to Cnidiria phylum, Anthozoa class, Scleractionia order, and Acroporidae family (Sims Citation2007). This species are widely distributed throughout the Indo-Pacific Ocean from East Africa to the Marshall Islands and Fiji (Dai and Horng Citation2009). The colonies are always found in shallow habitats (Veron Citation1995). It can be a dominant species of shallow mud reef flats. It was also found to be a pioneer species of reef flats during natural reef recovery after catastrophic destruction (Li et al. Citation2015). Montipora digitata colonies are digitated to arborescent with irregular, anastomosing branches. The branches become thinner, anastomosing, terete, or tapering from colonies in protected subtidal biotopes. Montipora digitata has a morphology similar to M. samarensis and M. altasepta, which can cause some difficulty in identifying them using only traditional morphological methods; thus, more up-to-date classification and molecular methods are urgently needed for coral identification.
Montipora digitata samples (TIOSOA-LCC-2013-Monti. Dig.) were collected in the Laboratory of Coral Conservation, Third Institute of Oceanography, SOA. Montipora digitata colony was obtained in 2013 from Houhai coral reefs in Sanya, Hainan Province. We used next generation sequencing (NGS) to perform low-coverage whole-genome sequencing according to our previous protocol (Shen et al. Citation2014). Initially, the raw NGS reads were generated from HiSeq X Ten (Illumina, San Diego, CA). About 0.09% of the raw reads (31,475 out of 33,430,528) were assembled de novo by using commercial software (Geneious V9, Auckland, New Zealand) in order to produce a single, circular form of a complete mitogenome with about an average 264× coverage.
The complete M. digitata mitogenome was 17,884 bp in size (GenBank KY094489), and its overall base composition was 24.9%, 14.2%, 24.2%, and 36.8% for A,C,G, and T, respectively, with a GC content of 38.3%. It showed a 99% identity to Anacropora matthai (GenBank AY903295) after the BLAST search against NCBI nr/nt database. rRNA and tRNA protein coding M. digitata mitogenome genes were predicted using DOGMA (Wyman et al. Citation2004), ARWEN (Laslett and Canback Citation2008), and MITOS (Bernt et al. Citation2013) tools followed by manual inspection. The complete mitogenome of M. digitata includes 13 unique protein-coding genes (PCGs), two ribosomal RNA (rrnL and rrnS) genes, and only three transfer RNA (trnW, trnR, and trnM) genes. All PCGs and tRNA and rRNA genes were encoded on the H-strand except for trnR, which is encoded on the L-strand. It is interesting to note one big intron (11,489 bp) is inserted in the ND5 gene.
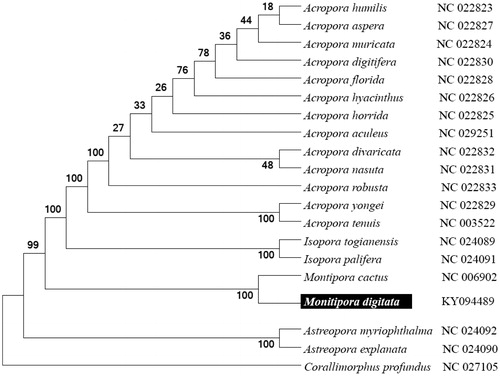
To validate the phylogenetic position of M. digitata, we used MEGA6 software (Tamura et al. Citation2013) to construct a maximum likelihood tree (with 500 bootstrap replicates and Kimura 2-parameter model) containing the complete mitogenomes for 20 Acroporidae family-derived species. Corallimorphidae-derived Corallimorphus profundus was used as an outgroup for tree rooting. Result shows M. digitata is close to M. cactus with high supported bootstrap value (). In conclusion, the complete M. digitata mitogenome deduced in this study provides essential and important DNA molecular data for further stony coral phylogenetic and evolutionary analysis.
Figure 1. Molecular phylogeny of M. digitata and related species based on complete mitogenome. The complete mitogenomes are downloaded from GenBank and the phylogenic tree is constructed by maximum likelihood method with 100 bootstrap replicates. The gene's accession number for tree construction is listed behind the species name.
Acknowledgements
Authors thank Dr. Chung-Der Hsiao from Chung Yuan Christian University for the help in genome sequencing.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Dai C, Horng S. 2009. Scleractinia Fauna of Taiwan: complex group. Taipei: National Taiwan University.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li YC, Han YS, Chen YQ, Xie HQ, Chen X, Zhou ZD. 2015. Damages of coral reef ecosystem by the giant clam excavation—a case study on Beijiao Reef of Xisha Islands. J Appl Oceanography. (In Chinese) 34:541–547.
- Shen KN, Yen TC, Chen CH, Li HY, Chen PL, Hsiao CD. 2014. Next generation sequencing yields the complete mitochondrial genome of the flathead mullet, Mugil cephalus cryptic species NWP2 (Teleostei: Mugilidae). Mitochondrial DNA. 27(3):1758–1759.
- Sims DW. 2007. Advances in marine biology. London: Elsevier Science.
- Tamura KG, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Veron JEN. 1995. Corals in space and time: the biogeography and evolution of the Scleractinia. Sydney: Comstock/Cornell.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
