Abstract
We analyzed the complete mitochondrial genome of the ‘Buckfast bee’, Apis mellifera, collected from North Ireland, UK. It consisted of a circular molecule of 16,353 bp. The genome contained 13 protein-coding, 22 tRNA, and 2 rRNA genes, along with one A + T-rich control region. The average AT content was 84.9%. The genes ATP8 and ATP6 shared 19 nucleotides. A phylogenetic analysis, suggested that the matriline ‘Buckfast bee’ has remained most closely related to the A. mellifera ligustica race from which it originated in 1917, despite being cross-bred with many other A. mellifera races over the past 100 years.
The Buckfast bee, Apis mellifera, was developed by Brother Adam at Buckfast Abbey, in South-West England, originally for their resistance towards the acarine mite, calmness on the comb and excellent honey production, all properties beekeepers value in A. mellifera (Adam Citation1987; Olszewski Citation2009; Holm Citation2014). In 1917, Brother Adam started breeding his new bee ‘Buckfast’ line with a few Italian queens (A. mellifera ligustica) (Adam Citation1987; Holm Citation2014). Over the following decades, he continually selected for traits he favored by crossing his ‘Buckfast line’ with a wide variety of bee races from across Europe (A. m. mellifera, A. m. ligustica, and A. m. cecropia), near Asia (A. m. anatoliaca), and Africa (A. m. sahariensis and A. m. monticola). Since his death in 1996, Buckfast Abbey and several honeybee breeder groups across Europe have continued to maintain the Buckfast strain on islands and other isolated places (Adam Citation1987; Holm Citation2014). Due to the large amount of cross-breeding with many different races, the ancestral lineage of the Buckfast bee remains unclear. Here, we analyzed the complete mitochondrial genome in order to help determine the maternal origin of the current ‘Buckfast bees’ maintained in Northern Island, UK.
An adult worker of a Buckfast bee queen previously crossed with an A. m. cecropia male in an apiary of Northern Ireland was collected in March 2017 (The specimen was stored in the National Museum of Nature and Science, Japan, accession number: NSMT-I-HYM 75326). Genomic DNA was extracted from its thoracic muscle tissue using a standard phenol/chloroform method. This was then sequenced using Illumina's MiSeq platform (Illumina, San Diego, CA). The resultant reads were assembled and annotated using the MITOS web server (Bernt et al. Citation2013) and Geneious R9 (Biomatters, Auckland, New Zealand). The phylogenetic tree was constructed using MEGA6 (Tamura et al. Citation2013) and TREEFINDER (Jobb Citation2015) using the nucleotide sequences of the 13 protein-coding genes (PCGs).
The mitochondrial genome of the ‘Buckfast bee’ consisted of a closed loop containing 16,353 bp (AP018432). That contained a heavy (H)-strand, which encoded nine PCGs and 14 tRNA genes, whereas the light (L)-strand encoded four PCGs, eight tRNA, and two rRNA genes. The start codon was ATT for the six PCGs; ATG for four PCGs; ATA for COI and ND3, and ATC for ND2. The stop codon for all PCGs was TAA.
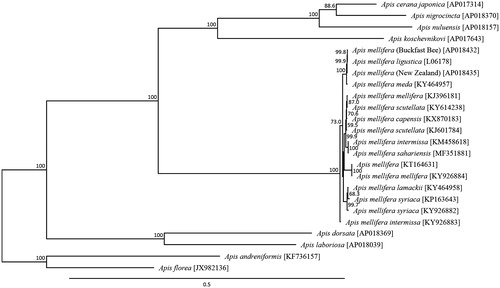
The A. mellifera haplotype of the non-coding region between tRNA-Leu and COII genes was the C lineage sequence, which was consistent with the findings of previous studies based on A. m. ligustica (Cornuet and Garnery Citation1991; Franck et al. Citation2000; Dall’Olio et al. Citation2007). A phylogenetic analysis was constructed using 13 mitochondrial PCGs across 17 Apis species (). This phylogenetic analysis suggested that the maternal line of the ‘Buckfast bee’ in Northern Ireland is most closely related to A. m. ligustica reflecting to persistence of the original Italian matrilines from which the Buckfast bee was originally derived from in 1917, over 100 years ago (Adam Citation1987; Holm Citation2014). The genetic distance between the ‘Buckfast bee’ and A. m. ligustica mitochondrial genome was 0.00036, which corresponds well to the genetic distance generally observed within A. mellifera subspecies.
Figure 1. Phylogenetic relationships (maximum likelihood) of the honey bees (Apis spp) and Apis mellifera races based on the nucleotide sequence of 13 protein-coding genes of the mitochondrial genome (Crozier and Crozier Citation1993; Gibson and Hunt Citation2015; Hu et al. Citation2015; Haddad Citation2015; Eimanifar et al. Citation2016a, Citation2016b; Eimanifar et al. Citation2017a, 2017Citationb, Citation2017c; Haddad et al. Citation2017; Nakagawa et al. Citation2017). The numbers at the nodes indicate bootstrap support inferred from 1000 bootstrap replicates. Alphanumeric terms indicate the GenBank accession numbers.
Acknowledgements
We are grateful to Mr. T. Wakamiya for valuable comments on the manuscript. This work was supported by a Japan public-private partnership student study abroad program TOBITATE! young ambassador program.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Adam B. 1987. Beekeeping at Buckfast Abbey with a section on mead making. West Yorkshire (UK): Northern Bee Books.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phyl Evol. 69:313–319.
- Cornuet JM, Garnery L. 1991. Mitochondrial DNA variability in honeybees and its phylogeographic implications. Apidologie. 22:627–642.
- Crozier RH, Crozier YC. 1993. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 133:97–117.
- Dall’Olio R, Marino A, Lodesani M, Moritz RFA. 2007. Genetic characterization of Italian honeybees, Apis mellifera ligustica, based on microsatellite DNA polymorphisms. Apidologie. 38:207–217.
- Eimanifar A, Kimball RT, Braun EL, Ellis JD. 2016a. The complete mitochondrial genome of the Cape honey bee, Apis mellifera capensis Esch. (Insecta: hymenoptera: apidae). Mitochondrial DNA Part B. 1:817–819.
- Eimanifar A, Kimball RT, Braun EL, Ellis JD. 2016b. The complete mitochondrial genome of the hybrid honey bee, Apis mellifera capensis × Apis mellifera scutellata, from South Africa. Mitochondrial DNA Part B. 1:856–857.
- Eimanifar A, Kimball RT, Braun EL, Fuchs S, Grünewald B, Ellis JD. 2017a. The complete mitochondrial genome of Apis mellifera meda (Insecta: Hymenoptera: Apidae). Mitochondrial DNA Part B. 2:268–269.
- Eimanifar A, Kimball RT, Braun EL, Moustafa DM, Haddad N, Fuchs S, Grünewald B, Ellis JD. 2017b. The complete mitochondrial genome of the Egyptian honey bee, Apis mellifera lamarckii (Insecta: Hymenoptera: Apidae). Mitochondrial DNA Part B. 2:270–272.
- Eimanifar A, Kimball RT, Braun EL, Fuchs S, Grünewald B, Ellis JD. 2017c. The complete mitochondrial genome of an east African honey bee, Apis mellifera monticola Smith (Insecta: Hymenoptera: Apidae). Mitochondrial DNA Part B. 2:589–590.
- Franck P, Garnery L, Celebrano G, Solignac M, Cornuet J-M. 2000. Hybrid origin of honeybees from Italy (Apis mellifera ligustica) and Sicily (A. m. sicula). Mol Ecol. 9:907–921.
- Gibson JD, Hunt GJ. 2015. The complete mitochondrial genome of the invasive Africanized honey bee, Apis mellifera scutellata (Insecta: Hymenoptera: Apidae). Mitochondrial DNA. 27:561–562.
- Haddad NJ. 2015. Mitochondrial genome of the Levant region honey bee, Apis mellifera syriaca (Hymenoptera: Apidae). Mitochondrial DNA. 30:1–2.
- Haddad NJ, Adjlane N, Loucif-Ayad W, Dash A, Naganeeswaran S, Rajashekar B, Al-Nakkeb K, Sicheritz-Ponten T. 2017. Mitochondrial genome of the North African Sahara Honey bee, Apis mellifera sahariensis (Hymenoptera: Apidae). Mitochondrial DNA Part B. 2:548–549.
- Holm E. 2014. Queen Breeding and Genetics – How to get better bees. West Yorkshire (UK): Northern Bee Books.
- Hu P, Lu ZX, Haddad N, Noureddine A, Loucif-Ayad Q, Qang YZ, Zhao RB, Zhang AL, Guan X, Zhang HX, Niu H. 2015. Complete mitochondrial genome of the Algerian honeybee, Apis mellifera intermissa (Hymenoptera: Apidae). Mitochondrial DNA. 26:1–2.
- Jobb G. 2015. TREEFINDAR. [accessed 2015 October 1]. http://www.treefinder.de/index_treefinder.html
- Nakagawa I, Maeda M, Chikano M, Okuyama H, Murray R, Takahashi J. 2017. The complete mitochondrial genome of the yellow-colored honeybee Apis mellifera (Insecta: Hymenoptera: Apidae) of New Zealand. Mitochondrial DNA Part B. 3:66–67.
- Olszewski K. 2009. Assessment of production traits in the buckfast bee. J Apic Sci. 53:79–90.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
