Abstract
As an endangered species, Magnolia kobus is distributed in Jeju island in Korea with only about 500–1000 individuals. In this study, we presented a complete chloroplast genome of M. kobus which is 159,443 bp and has four sub-regions: 87,484 bp of large single copy and 18,783 bp of small single copy regions are separated by 26,588 bp of inverted repeat regions including 113 genes (79 unique genes, four rRNAs and 30 tRNAs). Phylogenetic analysis using chloroplast genomes showed that M. kobus is a sister of M. insignis and M. laevifolia clade.
The Magnoliaceae is characterized by undifferentiated perianth (except sect. Buergeria), numerous stamens and carpels that are spirally arranged on an elongated receptacle (Nooteboom Citation1985). Recent classification system of Magnoliaceae (Figlar and Nooteboom Citation2004) suggests that there are two subfamilies (Liriodendroideae and Magnolioideae) in the family. According to the recent phylogenetic study, 11 distinctive major clades have been recognized with several basal polytomies in the subgenus Magnolioideae (Kim and Suh Citation2013).
Magnolia kobus DC. is a common and world-wide ornamental garden tree, however, its natural distribution is restricted in Japan and Korea. Especially, as endangered species, it is distributed in Jeju island in Korea with only about 500–1000 individuals. M. kobus is included in section Buergeria, in recent classification system (Figlar and Nooteboom Citation2004) and characterized by differentiated tepals.
We collected M. kobus at the natural population in Halla mountain in Jeju island N33°25′24.95”, E126°36′32.61”). Voucher specimen is deposited in Sungshin University Herbarium (SWU: S. Kim 201137). Total DNA was extracted from fresh leaves of M. kobus by using the ExgeneTM Plant SV mini kit (GeneAll, Seoul, Korea). Genome sequencing was performed using the GAIIx system (Solexa/Illumina, San Diego, CA). The NGS reads were matched and filtered against the chloroplast (cp) genome of Liriodendron tulipifera (Cai et al. Citation2006). The de novo assembled sequences were generated through three different assemblers, respectively: NGS Cell (Ver. 3.1.0; CLCBio, Denmark), ABySS (Ver. 1.2.5; Simpson et al. Citation2009), and Velvet (Ver. 1.0.15; Zerbino and Birney Citation2008). In a bid to utilize the advantages of three assembly programs, we aligned all contigs generated from three assemblers against the L. tulipifera cp genome using Sequencer 4.9 (Gene Code Corporation, Ann Arbor, MI). The completed cp genomes were annotated using DOGMA (Wyman et al. Citation2004) and were mapped with GenomeVx (Conant and Wolfe Citation2008). The M. kobus cp genome sequences were submitted to the GenBank (JX280396).
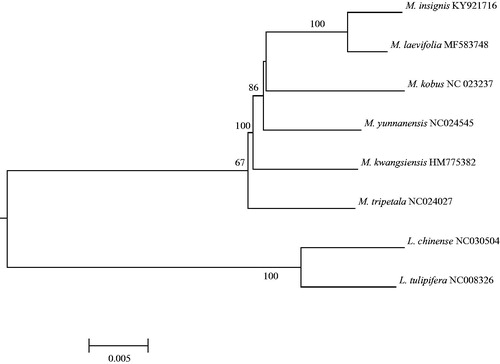
We generated around 24.5 Gbps sequences and obtained 122 Mbp short reads (ca. 760 X) as a filtered result. The cp genome of M. kobus is 159,443 bp and has four subregions: 87,484 bp of large single copy and 18,783 bp of small single copy regions are separated by 26,588 bp of inverted repeat regions. This cp genome includes 113 identified genes (79 unique genes, four rRNAs, and 30 tRNAs); 21 genes (six protein-coding genes, four rRNAs and 11 tRNAs) are duplicated in inverted repeat regions. The overall GC content is 39.28%. The phylogenetic tree was reconstructed with this cp genome and previously published cp genomes in Magnoliaceae (five in subgen. Magnolioideae and two in subgen. Liriodendroideae). Sequence alignment was conducted by MAFFT (Katoh and Standley Citation2013). The maximum likelihood (ML) analysis was performed using raxmlGUI version 1.3 (; Silvestro and Michalak Citation2012). Magnolia kobus is a sister of a clade comprising M. insignis and M. laevifolia. The cp genome resource will provide important information on the studies of M. kobus in Jeju island, which is an endangered species in Korea and on the future phylogenomic studies in the family.
Figure 1. Maximum likelihood tree based on “GTR + gamma + I” model using seven previously published chloroplast genome sequences in Magnoliaceae (Cai et al. Citation2006; Kuang et al. Citation2011; Yang et al. Citation2014; Li et al. Citation2016; Zhu et al. Citation2016; Xu et al. Citation2017; Zheng and Xu Citation2017) and that of M. kobus. The tree is rooted by subfamily Liriodendroideae. The numbers above the node indicate bootstrap value (500 replicates; > 50% are indicated).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, Boore JL, Jansen RK. 2006. Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol. 6:77.
- Conant GC, Wolfe KH. 2008. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 24:861–862.
- Figlar RB, Nooteboom HP. 2004. Notes on Magnoliaceae IV. Blumea. 49:87–100.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim S, Suh Y. 2013. Phylogeny of Magnoliaceae based on ten chloroplast DNA regions. J Plant Biol. 56:290–305.
- Kuang DY, Wu H, Wang YL, Gao LM, Zhang SZ, Lu L. 2011. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): implication for DNA barcoding and population genetics. Genome. 54:663–673.
- Li B, Li Y, Cai Q, Lin F, Meng Q, Zheng Y. 2016. The complete chloroplast genome of a Tertiary relict species Liriodendron chinense (Magnoliaceae). Conservation Genet Res. 8:279–281.
- Nooteboom H. 1985. Notes on Magnoliaceae. Blumea. 31:65–87.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Diver Evol. 12:335–337.
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res. 19:1117–1123.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Xu X, Zhang J, Zheng W. 2017. The complete chloroplast genome of threatened Magnolia laevifolia, a rare ornamental shrub with strong aromatic flowers. Conservation Genet Resour. https://doi.org/10.1007/s12686-017-0819-9.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Res. 14:1024–1031.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zheng W, Xu X. 2017. The complete chloroplast genome of endangered Manglietia insignis, a rare landscaping tree with red lotus-like flowers. Conservation Genet Resour. 10:27–30.
- Zhu A, Guo W, Gupta S, Fan W, Mower JP. 2016. Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol. 209:1747–1756.
