Abstract
The complete is mitochondrial genome of the Japanese bumblebee Bombus hypocrita sapporensis from Hokkaido Island, Japan is analysed using next generation sequencing. The mitochondrial genome of B. h. sapporensis was observed to be a circular molecule of 15,835 bp. The average AT content in the B. h. sapporensis mitochondrial genome was 85.53%. It was predicted to contain 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes, along with one A + T-rich control region. The PCGs had ATA, ATG, or ATT as the initiation codon and were terminated by the typical stop codon TAA, except for Cytb. All the tRNA genes typically formed a cloverleaf secondary structure, except for trnE, trnF, and trnS1. The molecular phylogenetic analysis indicated that the B. h. sapporensis from Hokkaido Island population was most similar to that of the geographically isolated B. h. sapporensis from Rebun Island.
The orange-tailed bumblebee in Far East Asian continent including North China was Bombus patagiatus, widely distributed to Russia, on the other hand, the orange-tailed bumblebee in Japan was true Bombus hypocrita (Williams, An, et al. Citation2012; Williams, Brown, et al. Citation2012). Williams, An, et al. (Citation2012) and Williams, Brown, et al. (Citation2012) indicated that orange-tailed bumblebee was a separate species in Japan and North China in mitochondrial DNA COI gene analyses using the DNA barcoding method. In morphological taxonomic analysis, Japanese B. hypocrita is classified into two subspecies, B. hypocrita sapporensis in Hokkaido Island, and B. h. hypocrita in Honshu, Shikoku and Kyushu Islands (Sakagami and Ishikawa Citation1969; Ito Citation1998; Ito and Munakata Citation1979), but the phylogenetic relationship of two B. hypocrita subspecies on Japan remains uncertain (Takahashi, Nishimoto, et al. Citation2016; Nishimoto et al. Citation2017; Takeuchi et al. Citation2017). Here we analysed the complete mitochondrial genome of orange-tailed bumblebee B. h. sapporensis from Hokkaido Island in Japan.
Adult workers of the B. h. sapporensis on wild flowers in the Otofuke Town (42°56′43″N 143°12′06″E), Hokkaido Islands, Japan, were collected in July 2017. The collected workers were transferred immediately to 99% ethanol for mitochondrial DNA analysis (the specimen was stored in the National Museum of Nature and Science, Japan, accession number: NSMT-I-HYM 74241). Genomic DNA isolated from males was sequenced using Illumina’s MiSeq platform (Illumina, San Diego, CA). The resultant reads were assembled and annotated using the MITOS web server (Bernt et al. Citation2013) and Geneious R9 (Biomatters, Auckland, New Zealand). Phylogenetic analysis was performed using the TREEFINDER (Jobb Citation2015) based on nucleotide sequences of the 13 protein-coding genes (PCGs).
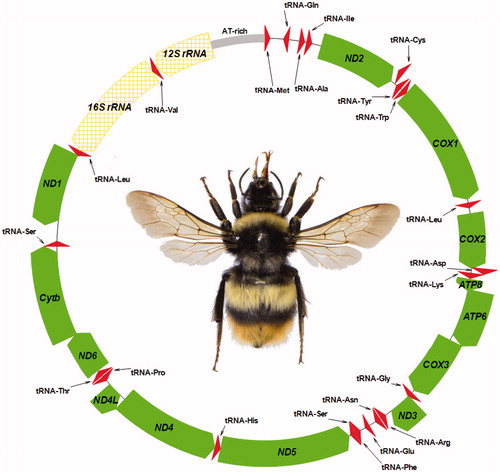
The average AT content of the B. h. sapporensis mitochondrial genome was 85.53%. Similar to the B. hypocrita mitochondrial genomes from Honshu and Rebun Islands (Takahashi, Nishimoto, et al. Citation2016; Nishimoto et al. Citation2017), the heavy strand (H-strand) was predicted to contain nine PCGs and 13 tRNA genes, and the light strand (L-strand) was predicted to contain PCGs, nine tRNA genes, and two rDNA genes (). The genes, ATP8 and ATP6, shared 19 nucleotides, ND4 and ND4L shared one nucleotide, and ND6 and Cytb shared 13 nucleotide. Six PCGs of the B. h. sapporensis mitochondrial genome started with ATA, ATP6, COIII, ND4, and Cytb started with ATG, and COII, ND5 and ND4L started with ATT; these starting codons have been found to be common in the other Bombus mitochondrial genomes (Cha et al. Citation2007; Hong et al. Citation2008; Du et al. Citation2015; Takahashi, Sasaki, et al. Citation2017; Zhao, Huang, et al. Citation2017; Zhao, Wu, et al. Citation2017). The stop codon in each of these PCGs was TAA, except for Cytb, which had TAG, as in other bumblebees.
Figure 1. Physical map of the mitochondrial genome of Bombus hypocrita sapporensis. Genes illustrated on the outside of the main circle are encoded on the heavy (H) strand; genes on the inside of the circle are encoded on the light (L) strand. The 13 protein-coding genes are labelled in filled arrow (green), 22 tRNA genes are labelled in triangle (red), and 16S rRNA and 12S rRNA genes are labelled in grid pattern arrow (yellow).
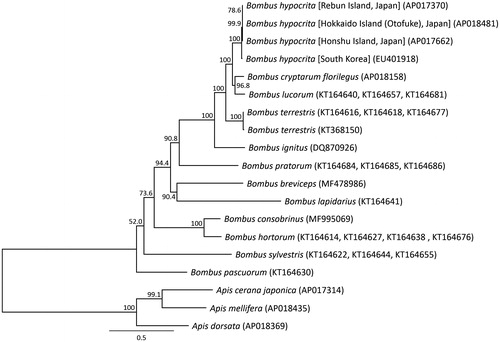
Molecular phylogenetic analysis was constructed by applying 13 mitochondrial PCGs with 12 closely related taxa (). The genetic distance and mutation site of the B. hypocrita in Japan taxa mitochondrial genomes were 0.0002–0.0018 and 2–20, on the other hand that of the orange-tailed bumblebee in Asia continent and the Japanese B. hypocrita were 0.0103–0.0107 and 113–117. The B. hypocrita from three Japanese Islands were shown to be a valid subspecies from the B. hypocrita in Asia continent by complete mitochondrial DNA sequence.
Figure 2. Phylogenetic relationships (maximum likelihood) among the species of the genus Bombus (Hymenoptera) based on the mitochondrial genome nucleotide sequence of the 13 protein-coding genes. Numbers beside the nodes are percentages of 1000 bootstrap values. The Apis cerana (Takahashi et al. Citation2016), Apis mellifera (Nakagawa et al. Citation2018), and Apis dorsata (Takahashi et al. Citation2017) were used as outgroup. Alphanumeric in parentheses indicates the DNA database of Japan accession numbers.
Acknowledgements
We thank Dr. Takuya Kiyoshi for helping our research.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo Metazoan Mitochondrial Genome Annotation. Mol Phylogenet Evol. 69:313–319.
- Cha SY, Yoon HJ, Lee EM, Yoon MH, Hwang JS, Jin BR, Han YS, Kim I. 2007. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene. 392:206–220.
- Du Q, Bi G, Zhao E, Yang J, Zhang Z, Liu G. 2015. Complete mitochondrial genome of Bombus terrestris (Hymenoptera: Apidae). Mitochondrial DNA. 5:1–2.
- Hong MY, Cha SY, Kim DY, Yoon HJ, Kim SR, Hwang JS, Kim KG, Han YS, Kim I. 2008. Presence of several tRNA-like sequences in the mitochondrial genome the bumblebee, Bombus hypocrita sapporoensis (Hymenoptera: Apidae). Genes Genomics. 30:307–318.
- Ito M, Munakata M. 1979. The bumblebees in Southern Hokkaido and Northwest Honshu, with notes on Blakiston Zoogeographical Line. J Low Temp Sci B. 37:81–105.
- Ito M. 1998. Species and distribution of Japanese Bumblebee. Insects Nat. 33:4–7 (in Japanese).
- Jobb G. 2015. TREEFINDER version of March 2011, Munich. http://www.treefnder.de.
- Nakagawa M, Maeda M, Chikano M, Okuyama H, Murray R, Takahashi J. 2018. The complete mitochondrial genome of the yellow coloured honeybee Apis mellifera (Insecta: Hymenoptera: Apidae) of New Zealand. Mitochondrial DNA B. 3:66–67.
- Nishimoto M, Okuyama H, Kiyoshi T, Nomura T, Takahashi J. 2017. Complete mitochondrial genome of the Japanese bumblebee, Bombus hypocrita hypocrita (Insecta: Hymenoptera: Apidae). Mitochondrial DNA B. 2:19–20.
- Sakagami SF, Ishikawa R. 1969. Note pre ´ liminaire sur la re ´partition ge ´ographique des bourdons japonais, avec descriptions et remarques sur quelques formes nouvelles ou peu connues. J Faculty Sci Hokkaido Univ (Zool). 71:152–196.
- Takahashi J, Deowanish S, Okuyama H. 2017. Analysis of the complete mitochondrial genome of the giant honeybee, Apis dorsata, (Hymenoptera: Apidae) in Thailand. Conserv Genet Resour. doi 10.1007/s12686-017-0942-7
- Takahashi J, Nishimoto M, Wakamiya T, Takahashi M, Kiyoshi K, Tsuchida K, Nomura T. 2016. Complete mitochondrial genome of the Japanese bumblebee, Bombus hypocrita sapporensis (Insecta: Hymenoptera: Apidae). Mitochondrial DNA B. 1:224–225.
- Takahashi J, Sasaki T, Nishimoto M, Okuyama H, Nomura T. 2017. Characterization of the complete sequence analysis of mitochondrial DNA of Japanese rare bumblebee species Bombus cryptarum florilegus. Conserv Genet Resour. doi:10.1007/s12686-017-0832-z
- Takahashi J, Wakamiya T, Kiyoshi T, Uchiyama H, Yajima S, Kimura K, Nomura T. 2016. The complete mitochondrial genome of the Japanese honeybee, Apis cerana japonica (Insecta: Hymenoptera: Apidae). Mitochondrial DNA B. 1:156–157.
- Takeuchi T, Takahashi M, Nishimoto M, Kiyoshi T, Tsuchida K, Nomura T, Takahashi J. 2018. Genetic structure of the bumble bee Bombus hypocrita sapporiensis, a potential domestic pollinator for crops in Japan. J Apic Res. 57:203–212.
- Williams PH, An J, Brown MJF, Carolan JC, Goulson D, Huang J, Ito M. 2012. Cryptic bumblebee species: consequences for conservation and the trade in greenhouse pollinators. PLoS One. 7:e32992.
- Williams PH, Brown MJF, Carolan JC, An J, Goulson D, Aytekin AM, Best LR, Byvaltsev AM, Cederberg B, Dawson R, et al. 2012. Unveiling cryptic species of the bumblebee subgenus Bombus s. str. worldwide with COI barcodes (Hymenoptera: Apidae). Syst Biodivers. 10:21–56.
- Zhao X, Huang J, Sun C, Jiandong A. 2017. Complete mitochondrial genome of Bombus consobrinus (Hymenoptera: Apidae). Mitochondrial DNA B. 2:770–772.
- Zhao X, Wu Z, Huang J, Liang C, An J, Sun C. 2017. Complete mitochondrial genome of Bombus breviceps (Hymenoptera: Apidae). Mitochondrial DNA B. 2:604–606.
