Abstract
The axillary seabream (Pagellus acarne, Risso 1827) belongs to the Sparidae family, order Perciformes. This high-valued commercial fish species is distributed along the northern and eastern Atlantic coasts from Norway to Senegal, and throughout the Mediterranean Sea. Its complete mitochondrial genome is 16,486 bp in length, consisting of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 2 non-coding regions (D-loop, 808 bp and L-origin, 29 bp). Its overall base composition is A: 26,8%, C: 29,0%, G: 17.6%, and T: 26.6%.
The axillary seabream (Pagellus acarne, Risso 1827) is a demersal fish belonging to the Sparidae family. Its geographical distribution extends from the Black Sea and the Mediterranean, along the west coast of Europe and Africa, to Norway and Angola (Santos et al. Citation1995). Pagellus acarne is a very common species especially in the Mediterranean, where it is prevalent in commercial and artisanal catches. It is listed as Least Concern in the IUNC Red List of Threatened Species, with the recommendation of reducing current fishing efforts (Russell et al. Citation2014). In this study, the complete mitochondrial genome of P. acarne was sequenced with Illumina HiSeq 2500 System (Illumina, San Diego, CA) (GenBank MG736083). The specimen was collected from the Mediterranean Sea (Gulf of Salerno, Tyrrhenian Sea; FAO area 37; N 40°34′47.40″', E 14°43′56.6'″) and identified based on morphological features. Total DNA was extracted from dorsal fin tissue and stored at the Department of Veterinary Medicine and Animal Production, University “Federico II” (Naples, Italy). The complete sequence was 16,486 bp in length, including 13 protein-coding genes, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA) and two non-coding regions (D-loop and L-origin). This gene arrangement is similar to the typical vertebrate mitogenome (Wang et al. Citation2008). Most of the genes were encoded on the heavy strand, while the NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes [Gln, Ala, Asn, Cys, Tyr, Ser (UCN), Glu, Pro] are encoded on the light strand. The nucleotide composition is A: 26,8%, C: 29%, G: 17.6%, and T: 26.6%, which is similar to other Sparidae mitogenomes (Xia et al. Citation2008; Shi et al. Citation2012; Dray et al. Citation2016). Protein-coding genes began with an ATG start codon, with the exception of COI and ND4 that start with GTG. Four types of stop codons revealed are TAA (ND1, ATP6, COIII, ND4L, ND5), AGG (COI), T (COII, ND4, Cytb) and TAG (ND2, ATP8, ND6). The 12S and 16S rRNA genes were located between the tRNAPhe (GAA) and tRNALeu (TAA) genes, and were separated by the tRNAVal gene as in other vertebrates (Li et al. Citation2016). The 22 tRNA genes vary from 66 to 73 bp in length. The 808 bp-long control region is located between tRNAPro (TGG) and tRNAPhe (GAA). The non-coding region (L-strand origin of replication) is 38 bp long and is located between tRNAAsn (GTT) and tRNACys (GCA).
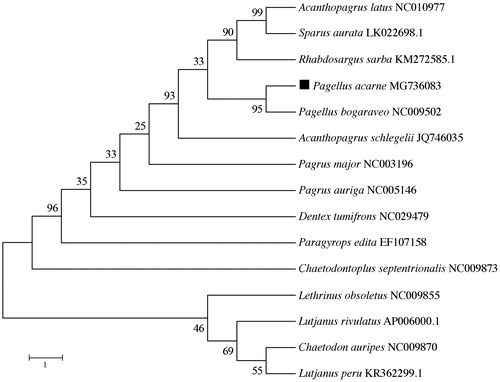
The phylogenetic position of P. acarne was validated using MEGA6 software (Tamura et al. Citation2013) to construct a maximum-likelihood (ML) tree with 1000 bootstrap replicates, containing the complete mitogenomes of 10 Sparidae species (). The resultant phylogeny shows that the P. acarne is closely related to P. bogaraveo with high bootstrap value supported. The study of the mitochondrial genome of P. acarne may reveal novel barcoding regions and deepen our knowledge of the evolution of sparid fishes.
Figure 1. The phylogenetic position of Pagellus acarne was validated by ML method with the complete mitogenomes of nine sparids and five arbitrary outgroup species (Lutjanus peru, Lutjanus rivulatus, Lethrinus obsoletus, Chaetodontoplus septentrionalis, Chaetodon auripes). Numbers above the nodes indicate 1000 bootstrap values. Mitogenome accession numbers are listed behind the species names.
Acknowledgements
The authors thank the Molecular Biology and Sequencing unit of the Stazione Zoologica Anton Dohrn for technical assistance.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Dray L, Neuhof M, Diamant A, Huchon D. 2016. The complete mitochondrial genome of the gilthead seabream Sparus aurata L. (Sparidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:781–782.
- Li J, Yang H, Xie Z, Yang X, Xiao L, Wang X, Li S, Chen M, Zhao H, Zhang Y. 2016. The complete mitochondrial genome of the Rhabdosargus sarba (Perciformes: Sparidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:1606–1607.
- Russell B, Carpenter KE, Pollard D. 2014. Pagellus acarne. The IUCN Red List of Threatened Species 2014: e.T170229A1297432.
- Santos MN, Costa Monteiro C, Erzini K. 1995. Aspects of the biology and gillnet selectivity of the axillary seabream (Pagellus acarne, Risso) and common pandora (Pagellus erythrinus, Linnaeus) from the Algarve (south Portugal). Fish Res. 23:223–236.
- Shi X, Su Y, Wang J, Ding S, Mao Y. 2012. Complete mitochondrial genome of black porgy Acanthopagrus schlegelii (Perciformes, Sparidae). Mitochondrial DNA. 23:310–312.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. 2008. Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): Insight into its phylogenic position within Cyprinidae. Gene. 424:96–101.
- Xia J, Xia K, Jiang S. 2008. Complete mitochondrial DNA sequence of the yellowfin seabream Acanthopagrus latus and a genomic comparison among closely related sparid species. Mitochondrial DNA. 19:385–393.
