Abstract
The common Dentex (Dentex dentex, Linnaeus 1758) has a significant economic importance and is a highly valued table fish in the Mediterranean region. The paucity of genetic information relating to sparids, despite their growing economic value, provides the impetus for exploring the mitogenomics of this fish group. Here, we sequenced D. dentex complete mitochondrial genome. The sequence is comprised of 16,652 bp and consists of 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes and a 2 non-coding regions (D-loop and L-origin). The overall nucleotide composition is: 27.5% of A, 28.7% of C, 26.9% of T, and 16.9% of G.
The common Dentex (Dentex dentex, Linnaeus 1758) is one of the most commercially caught fish species in the Mediterranean Sea, very appreciated in European markets. It is a littoral and benthopelagic sparid distributed in the Eastern Atlantic Ocean and in the entire Mediterranean Sea (Bauchot and Hureau Citation1986). Despite its significant economic importance, genetic information regarding this species is limited. We report the complete mitochondrial genome of D. dentex (GenBank MG727892). A specimen was caught in the Mediterranean Sea (N 41°46′58.3″, E 16°26′06.7″) and identified based on morphological features. DNA was extracted from dorsal fin tissue and is currently stored at Department of Veterinary Medicine and Animal Production, University “Federico II”, Naples, Italy. The complete mitogenome of D. dentex has been obtained from high-throughput sequencing on whole mitochondrial DNA with Illumina HiSeq 2500 System (Illumina, San Diego, CA, USA). The complete sequence is 16,652 bp long, containing 13 protein-coding genes, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA) and two non-coding regions (D-loop and L-origin). Mitochondrial structure and gene organization are in agreement with the typical vertebrate mitogenome (Wang et al. Citation2008). The majority of mitochondrial genes were encoded on the heavy strand, with the NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes [Gln, Ala, Asn, Cys, Tyr, Ser(UCN), Glu, Pro] being encoded on the light strand. Base composition is 27.5% A, 28.7% C, 26.9% T, and 16.9% G, similar to other Sparidae mitochondrial genomes (Shi et al. Citation2012; Dray et al. Citation2016). All protein-coding genes started with an ATG start codon but COI, which started with GTG. Stop codons were of 4 types, i.e. TAA (ND1, ND2, ATP8, ATP6, COIII, ND4L, ND5, ND6), AGG (COI), T (COII, ND4, CYTB) and TAG (ND3). The 12S and 16S rRNA genes were located between the tRNAPhe (GAA) and tRNALeu (TAA) genes, and were separated by the tRNAVal gene as in other vertebrates (Li et al. Citation2016). The 22 tRNA genes vary from 66 to 74 bp in length. The 971 bp long control region is located between tRNAPro (TGG) and tRNAPhe (GAA). The non-coding region (L-strand origin of replication) is 39 bp long and is located between tRNAAsn (GTT) and tRNACys (GCA).
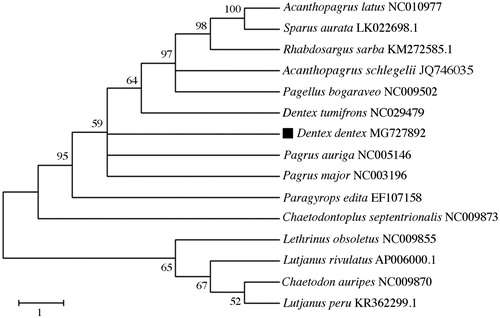
To validate the phylogenetic position of D. dentex, we used MEGA6 software (Tamura et al. Citation2013) with the entire mtDNA sequence of the sparids Acanthopagrus latus, A. schlegelii, D. tumifrons, Pagellus bogaraveo, Pagrus major, P. auriga, Parargyrops edita, Rhabdosargus sarba, and Sparus aurata. The species Lutjanus peru, L. rivulatus, Lethrinus obsoletus, Chaetodontoplus septentrionalis and Chaetodon auripes were used as outgroup for tree rooting (). The resultant phylogeny shows that D. dentex is closely related to D. tumifrons and Pagrus spp., in agreement with the work of Chiba et al. (Citation2009). The study of the mitochondrial genome of D. dentex and closely related species may reveal novel barcoding regions and inform on lineage diversification patterns in sparids.
Acknowledgements
The authors thank the Molecular Biology and Sequencing Unit of the Stazione Zoologica Anton Dohrn for technical assistance.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bauchot M-L, Hureau J-C. 1986. Sparidae. In: Whitehead PJP, Bauchot M-L, Hureau J-C, et al., editors. Fishes of the north-eastern Atlantic and the Mediterranean. Vol. 2. Paris: UNESCO. p. 883–907.
- Chiba SN, Iwatsuki Y, Yoshino T, Hanzawa N. 2009. Comprehensive phylogeny of the family Sparidae (Perciformes: Teleostei) inferred from mitochondrial gene analyses. Genes Genet Syst. 84:153–170.
- Dray L, Neuhof M, Diamant A, Huchon D. 2016. The complete mitochondrial genome of the gilthead seabream Sparus aurata L. (Sparidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:781–782.
- Li J, Yang H, Xie Z, Yang X, Xiao L, Wang X, Li S, Chen M, Zhao H, Zhang Y. 2016. The complete mitochondrial genome of the Rhabdosargus sarba (Perciformes: Sparidae). Mitochondrial DNA Part A. 27:1606–1607.
- Shi X, Su Y, Wang J, Ding S, Mao Y. 2012. Complete mitochondrial genome of black porgy Acanthopagrus schlegelii (Perciformes, Sparidae). Mitochondrial DNA. 23:310–312.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. 2008. Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): insight into its phylogenic position within Cyprinidae. Gene. 424:96–101.