Abstract
The complete mitochondrial genome sequence of black kite, Milvus migrans, one of the most common diurnal raptor, was characterized using next generation sequencing. The whole genome size was 18,016 bp and consisted of 13 protein-coding genes, 22 tRNAs, 2 rRNAs, a putative control region (CR), and a second control region (pseudo-CR). A frameshift mutation was found in the ND3 gene. Phylogenetic analysis illustrated monophyly of the subfamily Melieraxinae with high statistical support. The genetic resource obtained here will help to resolve taxonomic issues related to subspecies of M. migrans and will act as a starting point for conservation genetics.
The black kite, Milvus migrans, is a medium-sized bird of prey in the family Accipitridae and one of the most common diurnal raptors (Orta et al. Citation2017). It occurs throughout the Old World and Australasia (Sergio and Boto Citation1999). This species has experienced local declines in some regions, such as Portugal, Eastern Europe, and Russia, while remaining stable or increasing in Western Europe (Bustamante and Hiraldo Citation1990; Vinuela and Sunyer Citation1994; Sergio and Boto Citation1999). It is a winter migratory bird, although some individuals are considered year-round residents. The black kite is listed as a species of Least Concern (LC) on the International Union for the Conservation of Nature Red List of Threatened Species (BirdLife International Citation2016). However, in South Korea, this species has been classified as an endangered species II by the Ministry of Environment of Korea (NIBR Citation2011). In the present study, we sequenced and characterized the mitogenome of M. migrans.
An M. migrans sample (IN1594) was collected from Myongji-dong, Kangseo-gu, Busan, South Korea and deposited in the National Institute of Biological Resources at Inchoen, South Korea. Total genomic DNA was extracted from the muscle tissue using a DNeasy® Blood & Tissue Kit (Qiagen, Hilden, Germany). The complete mitogenome was determined using next generation sequencing (18,016 bp in length; GenBank MG930481). Annotation was performed with the online tool DOGMA (Wyman et al. Citation2004) and the software ARWEN (Laslett and Canbäck Citation2008). The complete mitogenome of M. migrans contained 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs, a putative control region (CR), and a second control region (pseudo-CR). The pseudo-CR (869 bp) was located between tRNA-Glu and tRNA-Phe. The PCGs were located on the H-strand, with the exception of ND and eight tRNA genes that were transcribed from the L-strand. The overall base composition of the mitogenome was 30.0% A, 32.5% C, 17.3% G, and 23.8% T; thus, the percentage of A and T (53.8%) was slightly higher than that of G and C (46.2%). Most of the protein-coding genes started with an ATG start codon, except COX I and ND5, which had GTG as an initiation codon. As is frequently reported for Accipitridae species (9 species of the 15 examined in this study, ), a frameshift mutation (insertion of “C”) in the ND3 gene was also found, which was located at position 174.
Figure 1. Phylogenetic tree of Acciptridae species based on the concatenated nucleotide sequences of the 13 protein-coding genes of their mitochondrial genomes. The analysis was performed using the MEGA 6.0 (Tamura et al. Citation2013) software, and bootstrap values are shown at the nodes. GenBank accession numbers for the sequences are indicated next to species designations. “$” indicates a species with a frameshift mutation in ND3.
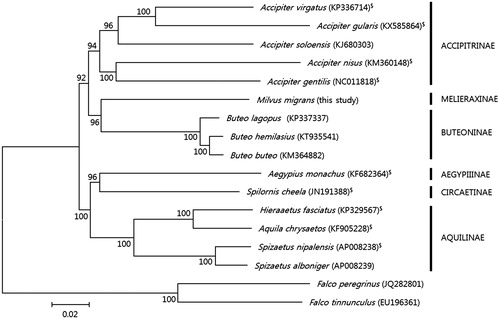
A phylogenetic analysis () performed using the 13 PCGs of the M. migrans mitogenome and additional available Accipitridae sequences retrieved from GenBank demonstrated the monophyly of the subfamily Melieraxinae with high statistical support. The Melieraxinae clustered closely to the Buteoninae, which is inconsistent with the results of Lerner and Mindell (Citation2005), which showed that Accipitrinae is closer to Melieraxinae. The complete mitogenome of M. migrans provides genetic resources that can be used to address existing taxonomic issues regarding the subspecies of M. migrans and that can be applied to conservation genetics.
Disclosure statement
The authors report no conflicts of interest and are alone responsible for the content and writing of the paper.
Additional information
Funding
References
- BirdLife International. 2016. Milvus migrans. The IUCN Red List of Threatened Species 2016: e.T22734972A95097654. [accessed 2017 Nov 27]. http://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T22734972A95097654.en.
- Bustamante J, Hiraldo F. 1990. Adoptions of fledglings by black and red kites. Animal Behav. 39:804–806.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Lerner HRL, Mindell DP. 2005. Phylogeny of eagles, old world vultures, and other accipitridae based on nuclear and mitochondrial DNA. Mol Phylogenet Evol. 37:327–346.
- National Institute of Biological Resources (NIBR). (2011). Red Data Book of Endangered Birds in Korea. Incheon, Korea: National Institute of Biological Resources (NIBR) (In Korean).
- Orta J, Marks JS, Garcia EFJ, Kirwan GM. 2017. Black Kite (Milvus migrans). In: del Hoyo, J, Elliott, A, Sargatal, J, Christie, DA, and de Juana, E, editors. Handbook of the Birds of the World Alive. Barcelona: Lynx Edicions. (Retrieved from https://www.hbw.com/node/52978 on 21 November 2017).
- Sergio F, Boto A. 1999. Nest dispersion, diet, and breeding success of Black Kites (Milvus migrans) in the Italian pre-Alps. J Raptor Res. 33:207–217.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molechular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 30:2725–2729.
- Vinuela J, Sunyer C. 1994. Black kite, Milvus migrans. In: GM Tucker and ME Heath, editors. Birds in Europe: their conservation status. BirdLife International (Conservation Series No. 3). Cambridge, UK: European Environment Agency. p. 148–149.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
