Abstract
Lycium ruthenicum is a precious desert plant with high medicinal and ecological values. It contains rich anthocyanins, which are safe natural pigments and could be used for natural food colorants. In this study, the complete chloroplast (cp) genome of L. ruthenicum was assembled on the basis of next generation sequencing. The whole genome is 154,996 bp in length, including two single copy regions (LSC: 85,993 bp and SSC: 18,213 bp) and two inverted repeat regions (IR: 25,395 bp). The overall G + C content of the genome is 37.9%. The cp genome consists of 111 unique genes, including 30 tRNA genes, 4 rRNA genes and 77 protein-coding genes. Phylogenetic analysis indicates that L. ruthenicum is closely clustered with Atropa belladonna, Przewalskia tangutica, and Scopolia parviflora.
The genus Lycium contains several most well-known medicinal and food plants in Solanaceae, such as L. barbarum, L. chinense, and Lycium ruthenicum. There are about 80 Lycium species around the world (Hitchcock Citation1932; Miller Citation2002; Levin and Miller Citation2005), with only seven species been found in China. Among them, L. ruthenicum is very important for the ecosystem because it is resistant to drought and salt stress and could reduce soil salinity (Zheng et al. Citation2011). It is also used to treat heart disease, ureteral stones, tinea, furuncle and gingival bleeding (Liu et al. Citation2014). Besides, the fruits of L. ruthenicum contain rich anthocyanins, which are safe natural pigments and could be used as food colorants. Recent studies have reported that the wild resources of L. ruthenicum rapidly declined due to human disturbance. Thus, it has been listed among the Key Protected Wild Plants (Category II) in Qinghai Province (http://www.qhys.gov.cn/html/42/202883.html). To scientifically and effectively conserve this plant, collecting more genetic information is important. Complete chloroplast (cp) genome could be applied to studies on population genetics and phylogenetics. It is useful to discuss the occurrence, evolution, and conservation of this precious endangered species.
Fresh and healthy leaves samples of wild L. ruthenicum were collected from the Yuepuhu County in Xinjiang Autonomous Region of China (39°12′39.6″N, 76°46′20.5″E,). Voucher herbarium specimens (GY17001) were deposited at the Herbarium of Xinjiang Normal University (XJNU). The total DNA was extracted using DNA Plantzol Reagent (Invitrogen, Shanghai, China) according to the manufacturer’s protocol, and then sequenced using Illumina HiSeqTM 2000 (San Diego, CA). The chloroplast sequences were assembled by NOVOPlasty 2.6.3 (Dierckxsens et al. Citation2017) with complete chloroplast sequence of Atropa belladonna (GenBank accession number: NC004561) as the reference. Finally, the whole sequence was annotated with Geneious 11.0.2 (Biomatters Ltd., Auckland, New Zealand). Then the complete circle was accomplished by OGDRAW (http://ogdraw.mpimp-golm.mpg.de/). The complete cpDNA sequence has been submitted to GenBank (MG976805). Lycium ruthenicum sequence was also aligned with the other Solanaceae cp genomes. The phylogenetic tree was constructed by MEGA6 (Tamura et al. Citation2013), with a Convolvulaceae species as the outgroup.
The whole genome of L. ruthenicum is 154,996 bp in length, including a large single copy region (LSC: 85,993 bp), a small single copy region (SSC: 18,213 bp) and two inverted repeat regions (IR: 25,395 bp). The overall G + C content of the genome is 37.9%. The cp genome consists of 111 unique genes, including 30 tRNA genes, 4 rRNA genes and 77 protein-coding genes. Seventeen genes are duplicated in the IR regions, including all four rRNA genes, seven tRNA genes and six protein-coding genes. Sixteen genes have one intron, while two genes have two introns. The ycf1, rps19 genes are crossing the SSC/IR and LSC/IR borders. The rps12 gene is trans-spliced, with the 5′ end located in the LSC and the 3′ end duplicated in the IR regions.
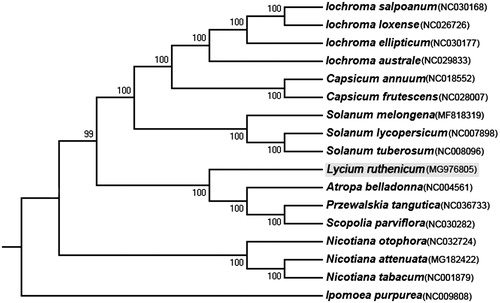
To figure out the phylogenetic position of L. ruthenicum within Solanaceae, we constructed neighbour joining (NJ) tree (with 1000 bootstrap replicates) using all Solanaceae species which have publicly available cp genome, with one species of Convolvulaceae (Ipomoea purpurea: NC009808) as the outgroup. It is found that L. ruthenicum is sister to a clade of Atropa belladonna, Przewalskia tangutica, and Scopolia parviflora (). In conclusion, this is the first research article of the complete cp genome of the L. ruthenicum, and this information which establish a solid foundation for future conservation genetic and phylogenetic studies.
Figure 1. Neighbour-joining phylogeny of Solanaceae based on 17 complete cp genomes. The accession numbers are listed as below: Iochroma salpoanum (NC030168), Iochroma loxense (NC026726), Iochroma ellipticum (NC030177), Iochroma australe (NC029833), Capsicum annuum (NC018552), Capsicum frutescens (NC028007), Solanum melongena (MF818319), Solanum lycopersicum (NC007898), Solanum tuberosum (NC008096), Lycium ruthenicum (MG976805), Atropa belladonna (NC004561), Przewalskia tangutica (NC036733), Scopolia parviflora (NC030282), Nicotiana otophora (NC032724), Nicotiana attenuata (MG182422), Nicotiana tabacum (NC001879), Ipomoea purpurea (NC009808).
Disclosure statement
The authors are really grateful to the genome data from public database. The authors report no conflicts of interest and are responsible for the content and writing of the paper.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Hitchcock CL. 1932. A monographic study of the genus Lycium of the western hemisphere. Ann Mo Bot Gard. 19:179–374.
- Levin RA, Miller JS. 2005. Relationships within tribe Lycieae (Solanaceae): paraphyly of Lycium and multiple origins of gender dimorphism. Am J Bot. 92:2044–2053.
- Liu Y, Zeng S, Sun W, Wu M, Hu W, Shen X, Wang Y. 2014. Comparative analysis of carotenoid accumulation in two goji (Lycium barbarum L. and L. ruthenicum Murr.) fruits. BMC Plant Biol. 14:269.
- Miller JS. 2002. Phylogenetic relationships and the evolution of gender dimorphism in Lycium (Solanaceae). Syst Bot. 27:416–428.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zheng J, Ding C, Wang L, Li G, Shi J, Li H, Wang H, Suo Y. 2011. Anthocyanins composition and antioxidant activity of wild Lycium ruthenicum Murr. from Qinghai-Tibet Plateau. Food Chem. 126:859–865.
