Abstract
The dragonfly Macromia daimoji Okumura, 1949 (Odonata: Macromiidae) has been listed as an Endangered insect in South Korea. We sequenced the complete 15,198 bp mitochondrial genome (mitogenome) of this organism, which is the first mitogenome sequence reported from the family Macromiidae. The genome includes a typical set of genes [13 protein-coding genes (PCGs), 2 rRNA genes, and 22 tRNA genes) and one non-coding region with an arrangement identical to that observed in most insect genomes. Phylogenetic analyses using concatenated sequences of the 13 PCGs and 2 rRNA genes using the Bayesian inference (BI) method placed Macromiidae, represented by M. daimoji, as a sister group to Libellulidae with the highest nodal support [Bayesian posterior probabilities (BPP) = 1]. Unlike conventional phylogenetic analysis, the suborders Anisozygoptera and Zygoptera formed a strong sister group (BPP =1), justifying the use of different molecular markers for phylogenetic analysis.
Macromia daimoji Okumura, 1949 (Odonata: Macromiidae), which is listed as an Endangered species in South Korea, is distributed in mid-Northern South Korea, Japan, and Southern Russia (Jeong Citation2012; http://www.me.go.kr/home/web/main.do). In Korea, limited ecological information for this species is available (Jeong Citation2012).
An adult male M. daimoji was collected from Yeongcheon gun, Gwangwon-do Province (38° 5' 47.20'' N, 127° 4' 29.40'' E), South Korea in 2009. This voucher specimen was deposited at the Chonnam National University, Gwangju, Korea, under the accession no. CNU7046. Using DNA extracted from the hind legs, four long overlapping fragments (LFs; COI-ND5, ND5-CytB, CytB-srRNA, and srRNA-COI) were amplified using four sets of primers designed from the available mitogenomes of Odonata (Lee et al. Citation2009; Wang et al. Citation2015; Yu et al. Citation2016; Jeong et al. Citation2017). Subsequently, these LFs were used as templates for amplifying 24 short fragments. The sequence data has been deposited in GenBank under the accession number MF990748.
We performed phylogenetic analysis using the concatenated nucleotide sequences of 13 protein-coding genes (PCGs) and 2 rRNA genes of 24 mitogenome sequences from Odonata. An optimal partitioning scheme (6 partitions) and substitution model (GTR + Gamma + I) were determined using PartitionFinder 2 and the Greedy algorithm (Lanfear et al. Citation2012; Lanfear, Calcott, Kainer, et al. Citation2014; Lanfear, Frandsen, et al. Citation2016). Bayesian inference (BI) analysis was conducted using Mr. Bayes ver. 3.2.2 (Ronquist et al. Citation2012) implemented on the CIPRES Portal ver. 3.1 (Miller et al. Citation2010).
The complete 15,198 bp mitogenome of M. daimoji was composed of 2 rRNAs, 22 tRNAs, 13 PCGs, and 1 major non-coding region referred to as the A + T-rich region (467 bp). The arrangement of this genome was identical to that typically observed for other insects (Cameron Citation2014). The A/T content of the whole mitogenome was 73.5%; however, it varied among the genes as follows: 86.4%, A + T-rich region; 75.5%, lrRNAs; 75.2%, srRNAs; 74.2%, tRNAs; and 72.4%, PCGs. Twelve PCGs had the typical ATN start codon, whereas ND1 had the atypical TTG codon. Ten of the 13 PCGs had a complete stop codon; however, COI, COII, and ND5 had an incomplete stop codon, T.
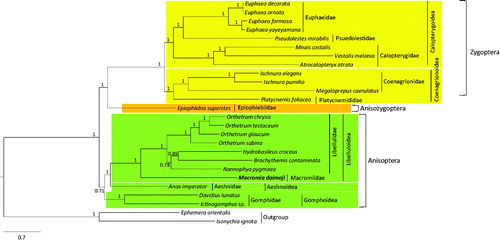
Results of phylogenetic analysis using the BI algorithm indicated sister relationship between Libellulidae and the newly added family, Macromiidae, with a strong nodal support (; BPP = 1), as has been previously shown using COI, 16S rRNA, 28S rRNA, and EF1-α sequences (Kim et al. Citation2014). Zygoptera was monophyletic with the highest nodal support (BPP = 1), and all the zygopteran superfamilies and families represented by more than one species were consistently and strongly supported as monophyletic groups. On the other hand, monophyletic Anisoptera was only moderately supported (BPP = 0.71), whereas the sister relationship between Aeshnoidea and Libelluloidea was also strongly supported (BPP = 1). The sister relationship between Zygoptera and Anisozygoptera (BPP = 1) was unconventional (Rehn Citation2003; Davis et al. Citation2011; Kim et al. Citation2014), but recent mitogenome-based phylogenetic results consistently supported the sister relationship between these two suborders (Yong et al. Citation2016; Jeong et al. Citation2017). We believe that in future, more species representing diverse taxonomic groups will help in understanding the odonate phylogeny.
Figure 1. Bayesian inference (BI) method-based phylogenetic tree for order Odonata obtained using concatenated sequences of 13 PCGs and 2 rRNAs. The numbers at each node indicate Bayesian posterior probabilities (BPP). The scale bar indicates the number of substitutions per site. Two species belonging to order Ephemeroptera were used as outgroups. GenBank accession numbers are as follows: B. contaminata, KM658172 (Yu et al. Citation2016); H. croceus, KM244659 (Tang et al. Citation2014); N. pygmaea, KY402222 (Jeong et al. Citation2017); O. chrysis, KU361233 (Yong et al. Citation2016); O. glaucum, KU361232 (Yong et al. Citation2016); O. Sabina, KU361234 (Yong et al. Citation2016); O. testaceum, KU361235 (Yong et al. Citation2016); Ictinogomphus sp., KM244673 (Tang et al. Citation2014); D. lunatus, EU591677 (Lee et al. Citation2009); A. imperator, KX161841 (Herzog et al. Citation2016); P. mirabilis, FJ606784 (unpublished); E. formosa, HM126547 (Lin et al. Citation2010); E. ornata, KF718295 (unpublished); E. decorata, KF718294 (unpublished); E. yayeyamana, KF718293 (unpublished); V. melania, JX050224 (Chen et al. Citation2015); A. atrata, KP233805 (unpublished); M. costalis, KU871065 (Lorenzo-Carballa et al. Citation2016); I. pumilio, KC878732 (Lorenzo-Carballa et al. Citation2014); I. elegans, KU958378 (Feindt et al. Citation2016a); P. foliacea, KP233804 (unpublished); M. caerulatus, KU958377 (Feindt et al. Citation2016b); E. superstes, JX050223 (Wang et al. Citation2015); E. orientalis, EU591678 (Lee et al. Citation2009); and I. ignota, HM143892 (unpublished).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117.
- Chen M-Y, Chaw S-M, Wang J-F, Villanueva RJT, Nuňeza OM, Lin C-P. 2015. Mitochondrial genome of a flashwing demoiselle, Vestalis melania from the Philippine Archipelago. Mitochondrial DNA. 26:720–721.
- Davis RB, Nicholson DB, Saunders ELR, Mayhew PJ. 2011. Fossil gaps inferred from phylogenies alter the apparent nature of diversification in dragonflies and their relatives. BMC Evol Biol. 11:252
- Feindt W, Herzog R, Osigus H-J, Schierwater B, Hadrys H. 2016a. Short read sequencing assembly revealed the complete mitochondrial genome of Ischnura elegans Vander Linden, 1820 (Odonata: Zygoptera). Mitochondrial DNA Part B. 1:574–576.
- Feindt W, Osigus H-J, Herzog R, Mason CE, Hadrys H. 2016b. The complete mitochondrial genome of the neotropical helicopter damselfly Megaloprepus caerulatus (Odonata: Zygoptera) assembled from next generation sequencing data. Mitochondrial DNA Part B. 1:497–499.
- Herzog R, Osigus HJ, Feindt W, Schierwater B, Hadrys H. 2016. The complete mitochondrial genome of the emperor dragonfly Anax imperator LEACH, 1815 (Odonata: Aeshnidae) via NGS sequencing. Mitochondrial DNA Part B. 21:783–786.
- Jeong KS. 2012. The dragonflies and damselflies of Korea. Seoul: Nature and Ecology.
- Jeong SY, Kim MJ, Wang AR, Kim S-S, An J, Kim I. 2017. Complete mitochondrial genome sequence of the tiny dragonfly, Nannophya pygmaea (Odonata: Libellulidae). Conservation Genet. Resour. (In Press) https://doi.org/10.1007/s12686-017-0823-0
- Kim MJ, Jung KS, Park NS, Wan X, Kim K-G, Jun J, Yoon TJ, Bae YJ, Lee SM, Kim I. 2014. Molecular phylogeny of the higher taxa of Odonata (Insecta) inferred from COI, 16S rRNA, 28S rRNA, and EF1-α sequences. Entomol Res. 44:65–79.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 14:82
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773.
- Lee EM, Hong MY, Kim MI, Kim MJ, Park HC, Kim KY, Lee IH, Bae CH, Jin BR, Kim I. 2009. The complete mitogenome sequences of the palaeopteran insects Ephemera orientalis (Ephemeroptera: Ephemeridae) and Davidius lunatus (Odonata: Gomphidae). Genome. 52:810–817.
- Lin CP, Chen MY, Huang JP. 2010. The complete mitochondrial genome and phylogenomics of a damselfly, Euphaea formosa support a basal Odonata within the Pterygota. Gene. 468:20–29.
- Lorenzo-Carballa MO, Thompson DJ, Cordero-Rivera A, Watts PC. 2014. Next generation sequencing yields the complete mitochondrial genome of the scarce blue-tailed damselfly, Ischnura pumilio. Mitochondrial DNA. 25:247–248.
- Lorenzo-Carballa MO, Tsubaki Y, Plaistow SJ, Watts PC. 2016. The complete mitochondrial genome of the broad-winged damselfly Mnais costalis Selys (Odonata: Calopterygidae) obtained by next-generation sequencing. Int J Odonatol. 19:191–198.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the 9th Gateway Computing Environments Workshop (GCE), New Orleans. p. 1–8.
- Rehn AC. 2003. Phylogenetic analysis of higher-level relationships of Odonata. Syst Entomol. 28:181–239.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mr. Bayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes-a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166
- Wang J-F, Chen M-Y, Chaw S-M, Morii Y, Yoshimura M, Sota T, Lin C-P. 2015. Complete mitochondrial genome of an enigmatic dragonfly, Epiophlebia superstes (Odonata, Epiophlebiidae). Mitochondrial DNA. 26:718–719.
- Yong H-S, Song S-L, Suana IW, Eamsobhana P, Lim P-E. 2016. Complete mitochondrial genome of Orthetrum dragonflies and molecular phylogeny of Odonata. Biochem Syst Ecol. 69:124–131.
- Yu P, Cheng X, Ma Y, Yu D, Zhang J. 2016. The complete mitochondrial genome of Brachythemis contaminata (Odonata: Libellulidae). Mitochondrial DNA Part A. 27:2272–2273.
