Abstract
The species tank goby (Glossogobius giuris) is assessed as Least Concern on the IUCN Red List of Threatened Species. In the present study, we first determined and described the complete mitochondrial genome of G. giuris. The circular genome is 16,595 bp in length. It has the typical vertebrate mitochondrial gene arrangement and consists of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and one non-coding control region. The overall base composition is 29.1% for A, 24.6% for T, 29.4% for C, and 17.0% for G. Phylogenetic analysis of 21 species in the family Gobiidae showed that G. giuris was clustered into the genus Glossogobius, subfamily Gobiinae. This information will contribute to future phylogenetic studies of Gobiidae and conservation strategies for G. giuris.
The species Glossogobius giuris is distributed throughout the Indo-west Pacific, and has been assessed as Least Concern on the IUCN red list (Kottelat Citation2013; Larson et al. Citation2016). So far, there are no known species-specific conservation measures in place for G. giuris (Larson et al. Citation2016). The mitogenome has been used as an effective tool for molecular taxonomy, species identification, and population genetic analyses in vertebrates (Galtier et al. Citation2009). However, the mitochondrial genetic information of G. giuris is unavailable. Therefore, the complete mitogenome of G. giuris was characterized and phylogenetically analysed in this study. We expect that the genomic data will provide essential information to genetic resources conservation and systematic study of G. giuris.
The specimen was collected from the Pearl River, Guangdong Province, China. Muscle was sampled and frozen in liquid nitrogen and then stored at −80 °C in South China Sea Fisheries Research Institute (Guangzhou, China). The mitochondrial DNA (mtDNA) was isolated using a Mitochondrial DNA Isolation Kit (Haling Biotech Shanghai, Co., Ltd., Shanghai, China). The whole mtDNA data were sequenced using the Illumina HiSeq Sequencing System (Illumina Inc., San Diego, CA). The clean data were acquired and assembled by the SOAPdenovo Assembler program and GapCloser (Beijing Genomics Institute, Shenzhen, China), and the assembled mtDNA was checked by PCR. The tRNA genes were identified using tRNAscan-SE 2.0 (Lowe and Chan Citation2016). A maximum likelihood tree of 21 Gobiidae species were constructed using MEGA6.06 software based on the complete mitochondrial genomes (Tamura et al. Citation2013).
The circular mitogenome of G. giuris (GenBank accession number NC_036674) is 16,596 bp in length. It has the typical vertebrate mitochondrial gene arrangement (Wang et al. Citation2017) and consists of 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and a control region (D-loop). Among these genes, ND6, tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser(UCA), tRNA-Glu, and tRNA-Pro are located on the light strand, while all of the remaining genes are located on the heavy strand. The overall base composition is A-29.1%, T-24.6%, C-29.4%, and G-17.0%. All of the PCGs initiate with ATG as start codon, except for COX1, which begins with GTG. This is quite common in vertebrate mtDNA (Yang et al. Citation2017). Six PCGs (ND1, ND4L, ND6, COX1, ATPase 6, and ATPase8) terminate with the typical TAA or TAG as stop codon, while ND5 ends with AGG. Five PCGs (ND2, COX2, COX3, ND3, and Cyt b) end with T––, and ND4 ends with TA–. Twenty-two tRNA genes, ranged in size from 68 to 75 bp, display a typical clover-leaf secondary structure, except for tRNA-Ser(AGC). The control region, with high A + T content (62.4%), is located between the tRNA-Pro and tRNA-Phe genes.
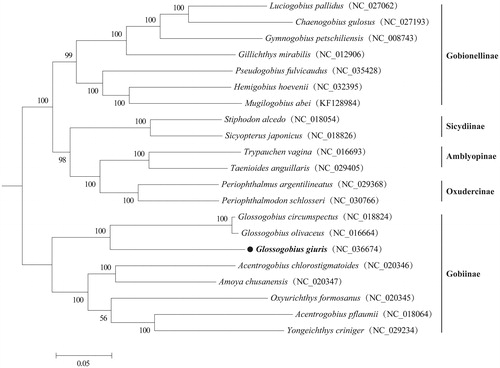
The phylogenetic tree constructed with 21 Gobiidae species (representing five subfamily) clearly demonstrated that G. giuris, as well as other Gobiinae species, was supported as a monophyletic group, and G. giuris was clustered in the clade of Glossogobius species (). In all, this genome will contribute to future phylogenetic studies of Gobiidae and conservation strategies for G. giuris.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Galtier N, Nabholz B, Glemin S, Hurst GDD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18:4541–4550.
- Kottelat M. 2013. The fishes of the inland waters of Southeast Asia: a catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. Raff Bull Zool. 27(Suppl.):1–663.
- Larson H, Britz R, Sparks JS. 2016. Glossogobius giuris. The IUCN Red List of Threatened Species (Least Concern ver 3.1). http://www.iucnredlist.org.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:54–57.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang PF, Chen SX, Ou YJ, Wen JF, Li JE. 2017. Complete mitochondrial genome of Toxabramis houdemeri (Cypriniformes; Cyprinidae) and its phylogenetic analysis. Mitochondrial DNA A DNA Mapp Seq Anal. 28:292–293.
- Yang QH, Lu Z, Zheng LY, Huang ZC, Lin Q, Wu JN, Zhou C. 2017. The complete mitochondrial genome of the Japanese seahorse, Hipppocampus Mohnikei (Syngnathiformes: Syngnathidae) and its phylogenetic implications. Conserv Genet Resour. 9:613–617.