Abstract
We assembled the plastome of the temperate, Southern Hemisphere liana Muehlenbeckia australis from high throughput sequencing data (paired-end Illumina reads) generated from total genomic DNA sequencing libraries. M. australis’ chloroplast genome sequence (GenBank: MG604297) is 163,484 bp in length and composed of long single copy (LSC; 88,166 bp) and short single copy (SSC; 13,486 bp) regions flanked by inverted repeats (IR; 30,916 bp each) typical for angiosperms. The plastome includes 131 genes comprising 83 protein-coding genes, 37 transfer RNA genes, eight ribosomal RNA genes, two possible pseudogenes, psbL and rpl23 with internal stop codons, and truncated repeats of ndhF and rps19 at IR boundaries.
Currently, seven species of Polygonaceae have published plastomes: four species of Fagopyrum Mill. (Logacheva et al. Citation2008; Cho et al. Citation2015; Wang et al. Citation2017) in tribe Fagopyreae, and two species of Rheum L. (Fan et al. Citation2015; Dagarova et al. Citation2017) and Oxyria sinensis Hemsl. (Luo et al. Citation2017) in the Rumiceae clade. These species have Northern Hemisphere distributions and an herbaceous habit. With Muehlenbeckia australis, we here contribute the first Southern Hemisphere, lianaceous woody species, and member of the Polygoneae clade to the pool of available plastomes.
We extracted DNA from ca. 20 mg of silica gel dried leaf material from a specimen growing in the living collection of the Royal Botanic Gardens Victoria (RBGV, 37°49′57.6″S 144°58′54.5″E; herbarium voucher for collection number TMS13-64 deposited at MEL) using the DNeasy Plant Mini Kit (Qiagen, Chadstone, Australia). A sequencing library was generated following the protocol of Schuster et al. (in press) and sequenced with an Illumina NextSeq 500 machine at The Walter and Eliza Hall Institute of Medical Research (WEHI) using an Illumina mid-output (2 × 150 Paired End) kit. Sequencing yielded 3,356,344 reads, which were de novo assembled with the software GENEIOUS v R10 (Kearse et al. Citation2012). Steps and parameters followed Gibbs’ (Citation2016) application note for chloroplast assembly. Annotations from Oxyria sinensis (NC_032031) were transferred to the final circular de novo plastome assembly and corrected manually, checking for start and stop codons, intron junctions, and inverted repeat (IR) boundaries.
The plastome of M. australis (GenBank: MG604297; ) is 163,484 bp long and has the typical four-part structure of angiosperms including two IR regions, each 30,916 bp in length, connecting the long single copy (LSC) region spanning 88,166 bp and a short single copy (SSC) region of 13,486 bp. In M. australis, the junction between LSC and IRB (JLB) lies within the rps19 gene, which is 279 bp long. Therefore, IRA includes 108 bp of a truncated inverted repeat of rps19 at JLA. The JSA junction between SSC and IRA lies in the ndhF gene, which is 2244 bp long, and therefore, IRB includes 62 bp of a truncated inverted repeat of ndhF at JSB. There are two possible pseudogenes, rpl23 and psbL, which have internal stop codons. However, Logacheva et al. (Citation2008) note that RNA may be edited to produce protein-coding copies, which could rescue functionality of these presumably essential genes involved in protein synthesis and photosynthesis.
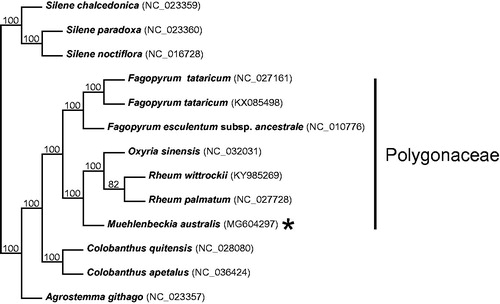
Figure 1. Cladogram resulting from a Neighbour-Joining (NJ) analysis including all currently available Polygonaceae plastomes and outgroup species (remainder of species not included in Polygonaceae clade indicated by a bar to the right of the tree). The alignment, NJ analysis, and tree rendering (Robinson et al. Citation2016) were generated on the MAFFT online server (Katoh et al. Citation2017), using alignment strategy FFT-NS-2 and leaving all other parameters at default settings. Bootstrap resampling = 100 with values shown on branches. Muehlenbeckia australis (MG604297) is indicated with an asterisk.
Sequencing of additional woody species of Polygonaceae will facilitate studies of shifts in substitution rates comparing herbaceous and woody species.
Acknowledgements
We thank Kate Roud, curator of New Zealand plants at the RBGV, for allowing us to collect material for DNA extraction and a herbarium voucher, Stephen Wilcox (WEHI) for generating sequence data, and Liz Milla (WEHI) for organizing a GENEIOUS workshop. GenBank accession number: MG604297. Specimen collection location: 37°49′57.6″S 144°58′54.5″E. Voucher specimen storage: The National Herbarium of Victoria, Melbourne (MEL [Index Herbariorum]; TMS13-64).
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Cho KS, Yun BK, Yoon YH, Hong SY, Mekapogu M, Kim KH, Yang TJ. 2015. Complete chloroplast genome sequence of Tartary buckwheat (Fagopyrum tataricum) and comparative analysis with common buckwheat (F. esculentum). PLoS One. 10:e0125332.
- Dagarova SS, Sitpayeva GT, Pak JH, Kim JS. 2017. The complete plastid genome sequence of Rheum wittrockii (Polygonaceae), endangered species of Kazakhstan. Mitochondrial DNA B. 2:516–517.
- Fan K, Sun XJ, Huang M, Wang XM. 2015. The complete chloroplast genome sequence of the medicinal plant Rheum palmatum L. (Polygonaceae). Mitochondrial DNA A. 27:2935–2936.
- Gibbs MD. 2016. Application note: de novo assembly and reconstruction of complete circular chloroplast genomes using Geneious, Biomatters Ltd. World Wide Web electronic publication; [accessed 2017 Aug 17]. https://assets.geneious.com/documentation/geneious/App+Note+-+De+Novo+Assembly+of+Chloroplasts.pdf.
- Katoh K, Rozewicki J, Yamada KD. 2017. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. DOI:10.1093/bib/bbx108.
- Kearse M, Moir R, Wilson A, Stone-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markovitz S, Duran C, et al. 2012. Geneius basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Logacheva MD, Samigullin TH, Dhingra A, Penin AA. 2008. Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp. ancestrale – a wild ancestor of cultivated buckwheat. BMC Plant Biol. 8:59.
- Luo X, Wang T, Hu H, Fan L, Wang Q, Hu Q. 2017. Characterization of the complete chloroplast genome of Oxyria sinensis. Conserv Genet Resour. 9:47–50.
- Robinson O, Dylus D, Dessimoz C. 2016. Phylo.io: interactive viewing and comparison of large phylogenetic trees on the web. Mol Biol Evol. 33:2183–2166.
- Schuster TM, Setaro SD, Tibbits JFG, Batty EL, Fowler RM, McLay TGB, Wilcox S, Ades PK, Bayly MJ. in review. Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae). PLoS One.
- Wang CL, Ding MQ, Zou CY, Zhu XM, Tang Y, Zhou ML, Shao JR. 2017. Comparative analysis of four buckwheat species based on morphology and complete chloroplast genome sequences. Sci Rep. 7:6514.
