Abstract
The mitochondrial genome of Sargassum yezoense (Yamada) Yoshida & T. Konno from Uljin, Korea was completely sequenced. This mitochondrial genome has 34,787 bp in length and consists of 65 genes including 37 protein-coding, three rRNA, and 25 tRNA genes. The overall GC content of the genome is 36.5%.
Sargassum C. Agardh (Sargassaceae, Phaeophyceae) is a widely distributed genus on rocky intertidal shores worldwide and represents one of the most species-rich genera of the marine macrophytes (Mattio and Payri Citation2011). Sargassum yezoense (Yamada) Yoshida & T. Konno is a large fucacean brown alga that is distributed in Japan (Agatsuma et al. Citation2002) and Korea (Oak and Lee Citation2006). This alga forms forests or meadows in sublittoral regions serving as nursery habitats and spawning grounds for marine invertebrates and fish (Fuse Citation1962). In this investigation, the complete mitogenome of S. yezoense was determined for the first time. This new information would contribute to the preservation of its genetic resources and provide better understanding of phylogenetic relationships of the Sargassum species and mitochondrial genome diversity in the Fucales.
We collected S. yezoense from Uljin, Gyeongsangbuk-do, Korea (36° 59′ 24.41″N 129° 24′ 56.62″E) on 5 April 2017. The specimen was deposited at the National Marine Biodiversity Institute of Korea (MABIK) under the accession number AL00070893. The total genomic DNA was extracted from fresh sample using an Exgene Plant SV Mini kit (GeneAll Biotechnology, Seoul, Korea) followed by preparation of a library based on the manufacturer’s instruction (Ion Torrent, Guilford, CT). Total genomic DNA was sequenced using the Ion Torrent Personal Genome Machine (Life Technologies, Carlsbad, CA), and high quality data, ca. 1.75 Gb were obtained. Total reads were assembled using the MIRA V. 4.0.2.1 and the CLC de novo assembler included in CLC Genomics Workbench V.5.5.1 (CLC Bio, Aarhus, Denmark). The size of the circular mitogenome produced is 34,767 bp (GenBank accession number MG674825) which is similar to the previously reported Sargassum mitogenomes (34,606–34,925 bp). The nucleotide composition is 26.9% A, 36.6% T, 21.5% G, and 15.1% C. The overall GC content is 36.6%. The S. yezoense mitogenome contains 65 genes, including 37 protein-coding, three rRNA, and 25 tRNA genes. The ATG start codon is used in all protein-coding genes. Twenty eight protein-coding genes end with TAA stop codon, five genes with TGA, and four genes with TAG, respectively. It was found that there are 10 cases of gene-overlapping ranging from 1 to 16 bp in size. All 25 tRNA genes ranged from 72 to 88 bp in length showing the typical cloverleaf secondary structures.
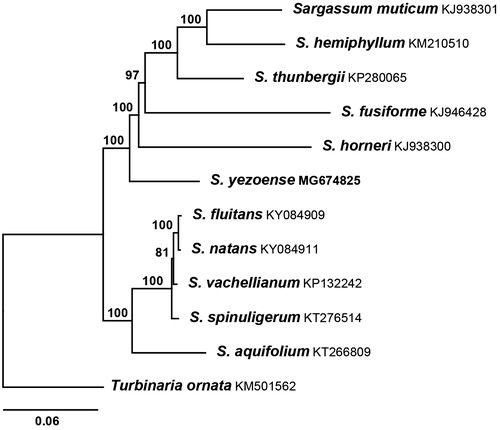
Phylogenetic analysis was carried out using 11 reported brown algal mitogenome sequences (Liu et al. Citation2015; Liu and Pang Citation2015, Citation2016a, Citation2016b; Bi and Zhou Citation2016; Liu, Pang, Chen Citation2016; Liu, Pang, Luo Citation2016; Amaral-Zettler et al. Citation2017). Sargassum yezoense combined tightly with S. muticum, S. hemiphyllum, S. thunbergii, S. fusiforme, and S. horneri with strong support values (). Based on the complete mitogenome of S. yezoense, new mt DNA marker could be selected and employed to study the phylogeography and phylogeny for Fucales.
Disclosure statement
The authors report no conflicts of interest. The authors are responsible for the content and writing of the paper.
Additional information
Funding
References
- Agatsuma Y, Narita K, Taniguchi K. 2002. Annual life cycle and productivity of the brown alga Sargassum yezoense off the coast of the Oshika Peninsula, Japan. Suisanzoshok. 50:25–30.
- Amaral-Zettler LA, Dragone NB, Schell J, Slikas B, Murphy LG, Morrall CE, Zettler ER. 2017. Comparative mitochondrial and chloroplast genomics of a genetically distinct form of Sargassum contributing to recent “Golden Tides” in the Western Atlantic. Ecol Evol. 7:516–525.
- Bi YH, Zhou ZG. 2016. Complete mitochondrial genome of the brown alga Sargassum vachellianum (Sargassaceae, Phaeophyceae). Mitochondrial DNA A. 27:2796–2797.
- Fuse S. 1962. The animal community in the Sargassum belt. Physiol Ecol. 11:23–45.
- Liu F, Pang SJ, Chen WZ. 2016. Complete mitochondrial genome of the brown alga Sargassum hemiphyllum (Sargassaceae, Phaeophyceae): comparative analyses. Mitochondrial DNA A. 27:1468–1470.
- Liu F, Pang SJ, Li X, Li J. 2015. Complete mitochondrial genome of the brown alga Sargassum horneri (Sargassaceae, Phaeophyceae): genome organization and phylogenetic analyses. J Appl Phycol. 27:469–478.
- Liu F, Pang SJ, Luo MB. 2016. Complete mitochondrial genome of the brown alga Sargassum fusiforme (Sargassaceae, Phaeophyceae): genome architecture and taxonomic consideration. Mitochondrial DNA A. 27:1158–1160.
- Liu F, Pang SJ. 2015. Mitochondrial genome of Turbinaria ornata (Sargassaceae, Phaeophyceae): comparative mitogenomics of brown algae. Curr Genet. 61:621–631.
- Liu F, Pang SJ. 2016a. Complete mitochondrial genome of the invasive brown alga Sargassum muticum (Sargassaceae, Phaeophyceae). Mitochondrial DNA A. 27:1129–1130.
- Liu F, Pang SJ. 2016b. Mitochondrial genome of Sargassum thunbergii: conservation and variability of mitogenomes within the subgenus Bactrophycus. Mitochondrial DNA A. 27:3186–3188.
- Mattio L, Payri CE. 2011. 190 years of Sargassum taxonomy, facing the advent of DNA phylogenies. Bot Rev. 77:31–70.
- Oak JH, Lee IK. 2006. Taxonomy of the Genus Sargassum (Fucales, Phaeophyceae) from Korea. Subgenus Bactrophycus section Halochloa and Repentia. Algae. 21:393–405.