Abstract
The common vole, Microtus arvalis belongs to the genus Microtus in the subfamily Arvicolinae. In this study, the complete mitochondrial genome of M. arvalis was recovered using shotgun sequencing and an iterative mapping approach using three related species. Phylogenetic analyses using the sequence of 21 arvicoline species place the common vole as a sister species to the East European vole (Microtus levis), but as opposed to previous results we find no support for the recognition of the genus Neodon within the subfamily Arvicolinae, as this is, as well as the genus Lasiopodomys, found within the Microtus genus.
The genus Microtus is one of the most diverse genera in the subfamily Arvicolinae. Consisting of about 70 different species it is one of the fastest radiating mammalian genera (Nowak Citation1999). The common vole (Microtus arvalis) is a small rodent widely distributed throughout Eurasia, ranging from the Atlantic coast in France to Central Russia. They experience a range of different climatic conditions from sea level to high altitude in the Alps (Fischer et al. Citation2011) and occupy a variety of different habitats such as farmland and grassland, making it a popular study species in ecological and evolutionary research. The species shows high levels of between population genetic differentiation (Heckel et al. Citation2005) and strong genetic clustering among populations on small scales (Schweizer et al. Citation2007). Although previous studies have used partial mitochondrial DNA sequences to show the presence of five main evolutionary lineages and to resolve their phylogenetic positioning within the genus Microtus (Fink et al. Citation2004), the complete mitochondrial genome of M. arvalis has not yet been published.
The male M. arvalis used in this study was sampled in 2015 outside the town of Lochow, Germany (52.690640°N, 12.455182°E) under permits from Landesumweltamt Brandenburg (RW-7.1 24.01.01.10). A voucher specimen was deposited at the University of Potsdam, Potsdam, Germany. DNA was extracted using the Qiagen DNeasy kit, built into an Illumina sequencing library and sequenced using an Illumina NextSeq 500. We assembled the full mitochondrial genome using an iterative mapping approach (Hahn et al. Citation2013) with three independent runs utilizing available Arvicolinae as reference bait sequences. Resultant sequences were aligned using Mafft v7.271 (Katoh and Standley Citation2013) and a final consensus sequence was built using Genious v9.0.5 (Kearse et al. Citation2012). We obtained a circular sequence, 16,286 bp in length (GenBank Accession No. MG948434) which was annotated using MITOS (Bernt et al. Citation2013). Finally, a phylogenetic analysis was performed on an alignment of our consensus sequence, all available complete mitochondrial arvicoline sequences and Cricetulus griseus. We produced a maximum-likelihood phylogenetic tree of all 13 protein-coding genes and tRNA genes, with an appropriate partitioning scheme and GTR + G as the substitution model as determined by PartitionFinder (Lanfear et al. Citation2012), with 1000 bootstrap replicates using RAxML-HPC2 on XSEDE v8.2.10 (Stamatakis Citation2014) on the Cipres server (Miller et al. Citation2010) ().
Figure 1. Maximum-likelihood tree of the subfamily Arvicolinae based on the sequences of 13 protein-coding genes and the tRNA genes of the mitochondrial genome using Cricetulus griseus as an outgroup (not shown here for graphical reasons). Bootstrap support values are shown at the branch nodes.
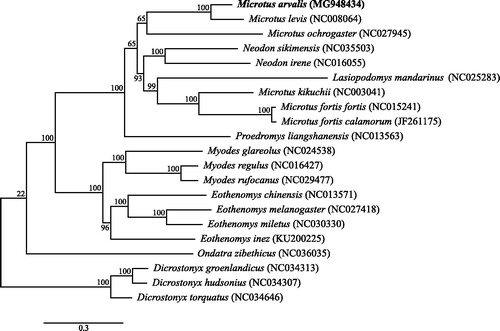
Phylogenetic analysis shows a well-supported sister-clade relationship between Arvicolini and Myodini. Furthermore, in concordance with other studies using mitochondrial or nuclear loci (Jaarola et al. Citation2004; Fink et al. Citation2010), we also find M. arvalis to be a sister species of Microtus levis. However, in contrast to Galewski et al. (Citation2006) who used the mitochondrial Cytb and nuclear Ghr genes, we found no support for the recognition of the genus Neodon, as this is found within the genus Microtus, as is the genus Lasiopodomys. Thus, mitogenomic analysis suggests that these genera should be subsumed within the Microtus genus. We hope publication of the mitochondrial genome of M. arvalis will help to understand the phylogenetic relationship within the Arvicolinae and the genus Microtus.
Disclosure statement
We would like to declare no conflict of interest. The authors are solely responsible for all content and writing.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Fink S, Excoffier L, Heckel G. 2004. Mitochondrial gene diversity in the common vole Microtus arvalis shaped by historical divergence and local adaptations. Mol Ecol. 13:3501–3514.
- Fink S, Fischer MC, Excoffier L, Heckel G. 2010. Genomic scans support repetitive continental colonization events during the rapid radiation of voles (Rodentia: Microtus): the utility of AFLPs versus mitochondrial and nuclear sequence markers. Syst Biol. 59:548–572.
- Fischer MC, Foll M, Excoffier L, Heckel G. 2011. Enhanced AFLP genome scans detect local adaptation in high-altitude populations of a small rodent (Microtus arvalis). Mol Ecol. 20:1450–1462.
- Galewski T, Tilak M, Sanchez S, Chevret P, Paradis E, Douzery EJ. 2006. The evolutionary radiation of Arvicolinae rodents (voles and lemmings): relative contribution of nuclear and mitochondrial DNA phylogenies. BMC Evol Biol. 6:80.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads - a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Heckel G, Burri R, Fink S, Desmet J-F, Excoffier L. 2005. Genetic structure and colonization processes in European populations of the common vole, Microtus arvalis. Evolution (NY). 59:2231–2242.
- Jaarola M, Martínková N, Gündüz İ, Brunhoff C, Zima J, Nadachowski A, Amori G, Bulatova NS, Chondropoulos B, Fraguedakis-Tsolis S, et al. 2004. Molecular phylogeny of the speciose vole genus Microtus (Arvicolinae, Rodentia) inferred from mitochondrial DNA sequences. Mol Phylogenet Evol. 33:647–663.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. In: 2010 Gatew Comput Environ Work. New Orleans (LA): IEEE. p. 1–8.
- Nowak R. 1999. Walker’s mammals of the world. Baltimore (MD): Johns Hopkins University Press.
- Schweizer M, Excoffier L, Heckel G. 2007. Fine-scale genetic structure and dispersal in the common vole (Microtus arvalis). Mol Ecol. 16:2463–2473.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
