Abstract
The Lucuma nervosa, native to Western Ghats of India, Malaysia and south-eastern Asia, is a tree member of the mulberry family (Sapotaceae). Chloroplast genome sequences play an significant role in the development of molecular markers in plant phylogenetic and population genetic studies. In this study, we report the complete chloroplast genome sequence of L. nervosa for the first time. The chloroplast genome is 157,920 bp long and includes 113 genes. Its LSC, SSC, and IR regions are 88,123, 18,861, and 25,468 bp long, respectively. Phylogenetic tree analysis exhibited that L. nervosa was clustered with other Sapotaceae species with high bootstrap values.
The Lucuma nervosa is a perennial evergreen fruit tree of the family Sapotaceae. It is native to the tropics of Cuba and South America, and are cultivated sporadically in Guangdong, Guangxi, southern Yunnan and Hainan of China (Liu et al. Citation2013). Its fruit can be eaten fresh or processed into fruit paste and cream with edible rate above 70%, which was rich in nutrition, including soluble solids 14.48–17.53%, sugar 29.1–30.5%, starch 5.6–8.1%, crude fat 1–1.4%, every 100 g flesh contains vitamin C 29.38–57.55 MG, acid content of 0.079–0.165% (Deng et al. Citation2013). Furthermore, the trees are beautiful and can be used for ornamental cultivation (Deng et al. Citation2013).
There was only one complete chloroplast genome reported in the family Sapotaceae (Jo et al. Citation2016). In this study, we report the complete chloroplast genome of L. nervosa, the second complete plastome sequence from the family Sapotaceae. DNA material was isolated from mature leaves of a L. nervosa plant cultivated in the plant garden of Yunnan Institute of Tropical Crops (YITC), Jinghong, China by using DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). A specimen of this tree was conserved in YITC. About 10 μg isolated DNA was sent to BGI, Shenzhen for library construction and genome sequencing on the Illumina Hiseq 2000 Platform. After genome sequencing, a total of 3.2 Gbp reads in fastq format were obtained and subjected to chloroplast genome assembly. The complete chloroplast genome was annotated with Dual Organelle GenoMe Annotator (DOGMA; Wyman et al. Citation2004) and submitted to the GenBank under the accession number of MH018545.
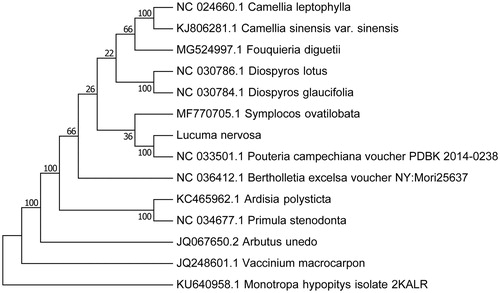
Our assembly of the L. nervosa resulted in a final sequence of 157,920 bp in length with no gap. The overall A–T content of the chloroplast genome was 61.2%. This chloroplast genome included a typical quadripartite structure with the large single copy (LSC), small single copy (SSC), and inverted repeat (IR) regions of 88,123, 18,861, and 25,468 bp long, respectively. Genome annotation showed 113 full length genes including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. The genome organization, gene content, and gene relative positions were almost identical to the previously reported Sapotaceae chloroplast genomes (Jo et al. Citation2016). To validate the phylogenetic relationships of L. nervosa in the Ericales, we constructed a maximum likelihood tree using 14 super-Ericales taxa. Phylogenetic analysis was performed on a data set that included 79 protein-coding genes and four rRNA genes from the 27 selected taxa using RAxML v. 7.7.1 (Stamatakis et al. Citation2008). The 83 gene sequences (82,565 bp in length) were aligned with the MAFFT (Katoh and Standley Citation2013). The resulting tree shows that L. nervosa forms a clade with the species of Ebenaceae and Primulaceae with a 100% bootstrap value (). The Sapotaceae represented by L. nervosa form a sister group to the Primulaceae–Ebenaceae clade within the Ericales. The close relationship of Sapotaceae to Lecythidaceae was suggested by a previous study (Anderberg et al. Citation2002).
Acknowledgements
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. The project was funded by Sci-Tech Innovation System Construction for Tropical Crops Grant of Yunnan Province (No. RF2017/RF2018).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Anderberg AA, Rydin C, Källersjö M. 2002. Phylogenetic relationships in the order Ericales sl: analyses of molecular data from five genes from the plastid and mitochondrial genomes. Am J Bot. 89:677–687.
- Deng CJ, Liu XM, Zhao SM, Qin LI, Zhang JC, Zhang GY, Chen WQ. 2013. Recent advances in research of tropical and sub-tropical fruit tree: Lucuma nervosa.Jinghong, PR China: Tropical Agricultural Science & Technology.
- Jo S, Kim HW, Kim YK, Cheon SH, Kim KJ. 2016. The first complete plastome sequence from the family Sapotaceae, Pouteria campechiana (Kunth) Baehni. Mitochondrial DNA B. 1:734–736.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu SF, Gu ZH, Zhong ML. 2013. On development situation and potential of Lucuma nervosa cultivation in China. J Anhui Agric Sci. 12:25.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20:3252–3255.