Abstract
Genus Prionailurus comprises four species, i.e. Leopard cat, Rusty spotted cat, Fishing cat, Flat-headed cat, listed under IUCN as threatened species except P. bengalensis. In U.S.A., P. bengalensis is listed as Endangered. Subspecies of P. bengalensis, i.e. iriomotensis is listed as Critically Endangered under IUCN since 2008. The present study describes the use of two markers 12SrRNA and cytochrome b genes to differentiate the three species and three subspecies of Prionailurus. Old taxidermy samples (three skin samples) of P. viverrinus and P.b. horsfieldi were used from India for the study. The study done by using DNAsp v5, MEGA 6.0, and Network 5.0.0.1, proved that both gene markers are useful for differentiating the species and subspecies of Prionailurus. This study is also the first study to present forensically informative nucleotide sequence (FINS) for three species and three subspecies of Prionailurus.
Introduction
The genus Prionailurus comprises of species Leopard cat (Prionailurus bengalensis), Rusty spotted cat (Prionailurus rubiginosus), Fishing cat (Prionailurus viverrinus), and Flat-headed cat (Prionailurus planiceps). Leopard cats are the most widely distributed Asian small cats (Nowell and Jackson Citation1996; Sunquist and Sunquist Citation2002). Their range with subspecies bengalensis, javanensis, sumatranus, chinensis, horsfieldi euptilurus/euptilura, borneoensis, trevelyani, alleni, iriomotensis, heaneyi, and rabori extends from the Amur region in the Russian Far East over the Korean Peninsula, China, Indochina, the Indian Subcontinent, to the West in northern Pakistan, and to the south in the Philippines and the Sunda islands of Indonesia (Ross et al. Citation2010). P.b. horsfieldi ranges in Kashmir, Punjab, Kumaon, Nepal, Sikkim, and Bhutan (Vella et al. Citation2002); Amur leopard cat P.b. euptilurus/euptilura is distributed in eastern Siberia, Manchuria, and Korea (Vella et al. Citation2002) and on the Tsushima Island (Werdelin and Olsson Citation2008). Iriomote cat P.b. iriomotensis is found exclusively on the tiny island of Iriomote in the Japanese Archipelago (Imaizumi Citation1967).
Leopard cat, Fishing cat (Endangered by IUCN, A2cd +4cd ver 3.1), Flat headed cat (Endangered C1 ver 3.1.) are commercially traded internationally for the fur trade and also killed in retribution (Nowell and Jackson Citation1996; Duckworth et al. Citation2009; Ross et al. Citation2010, http://www.iucnredlist.org).
Mitochondrial 12S rRNA and cytochrome b genes have been applied to understand the interspecies and intraspecies genetic diversity. In the present study, two skin samples were used belonging to P.b. horsfieldi and P. viverrinus for amplification of genes, i.e. 12SrRNA and cytochrome b and of other species and subspecies of the genus Prionailurus were used from NCBI. Since the gene sequences (12SrRNA and cytochrome b) of only two subspecies, i.e. P.b. iriomotensis and P.b. euptilurus are available at NCBI thus the present study is confined to three subspecies of P. bengalensis, i.e. P.b. horsfieldi, P.b. euptilurus, and P.b. iriomotensis. This study is a first attempt of molecular characterization of three species out of four known species of genus Prionailurus, i.e. P. bengalensis, P. viverrinus, and P. planiceps and (as no nucleotide database is available for P. rubiginosus) and four subspecies P.b. iriomotensis and P.b. euptilurus of P. bengalensis horsfieldi and P.b. bengalensis.
This study is useful in wildlife forensic for species identification and enforcement of law and regulations by providing strong scientific proofs and for their status based on forensically informative nucleotide sequencing (FINS) (Bartlett and Davidson Citation1992). The technique combines DNA sequencing and phylogenetic analysis. The author discussed the genetic difference between three subspecies of P.b. horsfieldi, P.b. iriomotensis, and P.b. euptilurus hence the origin of the seized wildlife parts and products.
Materials and methods
Sample collection
In this study, taxidermy skin samples were used. These taxidermy samples (0.5 cm × 0.5 cm) were collected from Mammals section of Zoological Survey of India, Kolkata (). Additionally, 12s rRNA and cytochrome b available sequences of small cats of genus Prionailurus were obtained from GenBank ().
Table 1. Sample details.
DNA isolation, PCR amplification, and sequencing
After washing with MilliQ water and cleaning, the skin samples were hydrated before digestion by incubating the dried skin sample for 24 h in 1 ml TE solution (Tris 10 mM and EDTA 1 mM, pH 7.6) (Barros and Morgante Citation2007). After 24 h of hydration, the DNA was isolated from skin sample using HiPur ATM Forensic Sample Genomic DNA Purification Kit (HIMEDIA).
12s rRNA sequences were amplified using a set of primer pair, L1091 and H1478, and a primer set of L14841 and H15149 was used to amplify cytochrome b gene (Kocher et al. Citation1989). The PCR reaction was performed in Q-cycler, Quanta Biotech (Surrey, UK), in a total volume of 25 µl of reaction mixture (10× PCR-with MgCl2, 2.5 µl; 10 mM dNTP’s, 2.5 µl; 5 pmol primer, 0.45 µl each; 15 ng of DNA template; 1.5 U Taq enzyme). Polymerase chain reaction consisted of initial denaturation of 94 °C for four minutes and each cycle of denaturation for 1 min at 94 °C, hybridization for 1 min at 55 °C (50 °C for cytochrome b) and extension for 1 min at 72 °C followed by final elongation for 10 min at 72 °C. The cycle was repeated for 35 times. The PCR products were sequenced using ABI's AmpliTaq FS dye terminator cycle sequencing chemistry on an automated ABI 3100 Genetic Analyser. All experiments were performed in a PCR workstation (Bangalore GeNeiTM). Negative controls were used in all DNA extraction and PCR amplification to control for potential contamination. 12SrRNA gene and cytochrome b gene sequences thus generated are submitted to NCBI after conducting sequence alignment by Bioedit and by checking their similarity with species of genus Prionailurus.
Data analysis
Sequences were visualized and edited using Chromas 1.6 (Technelysium Pty Ltd., South Brisbane, Australia). To crosscheck, quarry sequences were compared using GenBank BLAST (http://www.ncbi.nlm.nih.gov/BLAST). CLUSTAL W was used to compare DNA sequence data implemented in BioEdit v 7.0.9.0 software (Hall Citation1990) with outgroup Canis lupus lupus (AM711902.1). All sequences were proof read and analysed by using MEGA6.0 (Tamura et al. Citation2011) and were aligned by using ClustalW (Thompson et al. Citation1994). MEGA6.0 was used for finding the conserved, variable, parsimony informative and singleton sites as well as for phylogeny construction. Two methods were used for phylogeny construction: (i) maximum likelihood and (ii) neighbour joining with Kimura 2 Parameter (Pevsner Citation2009). All trees were subjected to bootstrap analysis with 1000 replicates to get bootstrap value support. The complete mitochondrial genome of Cunis lupus lupus was retrieved from NCBI database to be used as outgroup.
Results and discussion
12Sr RNA gene was amplified from extracted DNA of P. viverrinus, P.b. bengalensis, and P.b. horsfieldi. After sequencing, the unambiguous lengths of 12S rRNA (ca. 377 bp) were obtained and the same was tried for cytochrome b. The BLAST result indicated that the obtained sequences matched 99–100% with their respective species. In 12srRNA with 377 bp, the author observed five haplotypes, 312 conserved sites, 59 variable sites, 17 parsimony informative sites, and 41 singleton sites and in cytochrome b with 1100 bp, the author observed seven haplotypes; 816 conserved sites, 284 variable sites, 84 parsimony informative sites, and 200 singleton sites.
Species specific sites in 12S rRNA gene
In relation to the complete mitochondrial genome of Canis lupus lupus out of 1100 bp of 12SrRNA gene, there were 320, 320, 319, 321, and 301 conserved sites, 44, 44, 44, 43, and 58 variable sites and 49, 49, 47, 48, and 63 parsimony informative sites were observed in P.b. iriomotensis, P.b. euptilurus, P. bengalensis, P.b. horsfieldi, and P. viverrinus, respectively. Six singleton, one singleton, and 26 singleton sites were observed in P. bengalensis (D28889.1), P. bengalensis (D28890.1), and P. viverrinus (KM093869.1), respectively, and for P.b. iriomotensis, P.b. euptilurus, and P. bengalensis horsfieldi it was noted to be zero. The nucleotide composition of the entire sequence region of 12S rRNA gene (377 bp) is A 36.25%, T/U 24.39%, C 21.77%, and G 17.59%.
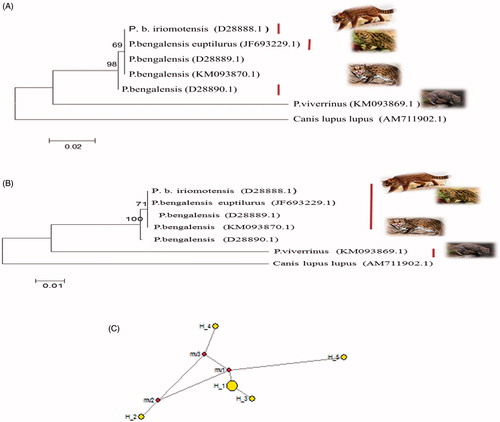
Concurrent evolutionary trees were obtained by using MEGA 6 with maximum likelihood and neighbour joining algorithms (), where P.b. iriomotensis and P.b. euptilurus, along with P. bengalensis (D28889) and P. bengalensis (D28890) and P.b. horsfieldi formed one clade. However, by using DnaSPv5 and network analysis by using NETWORK software indicated that haplotype 1 belongs to P. viverrinus and haplotype 2 is shared by P.b. iriomotensis, P.b. euptilurus, and P. bengalensis (D28889) indicating the common origin. Haplotype 3 belongs to P. bengalensis (D28890.1 with unknown subspecies) and haplotype 4 belongs to P.b. horsfieldi ().
Figure 1. 12SrRNA based ML tree topology (A) and NJ tree topology (B) of species and subspecies of Prionailurus. The evolutionary distances were computed using the Kimura two-parameter method and are in the units of the number of base substitutions per site. The analysis involved seven nucleotide sequences including outgroup Canis lupus lupus. Median-joining network by using 12SrRNA gene of genus Prionailurus indicating four haplotypes of species and subspecies of Prionailurus. Haplotype 5 belongs to outgroup (C).
Cytochrome b gene analysis
Genetic analysis was done by MEGA 6 with seven sequences, ca. 1100 bp with Canis lupus lupus as outgroup inferred 284 variable/polymorphic (segregating) sites and seven haplotypes.
In relation to the complete mitochondrial genome of Canis lupus lupus (AM711902.1) out of 1100 bp cytochrome b sequence as determined by MEGA 6, there were 868, 870, 865, 886, and 867 conserved sites; 232, 230, 233, 214, and 233 variable sites and 223, 231, 244, 216, and 232 parsimony informative sites in P.b. irimotensis, P.b. euptilurus, P. viverrinus, P. planiceps, and P. bengalensis, respectively. Singleton sites were 0 in P.b. irimotensis, two in P.b. euptilurus, 16 in P. viverrinus, 17 in P. planiceps, and two in P. bengalensis. These sites can be used to differentiate the species and subspecies of Prionailurus. Six haplotypes were present. Hap 1 for P.b. iriomotensis, Hap 2 for P.b. euptilurus, Hap3 and Hap 4 belongs to P. viverrinus, Hap 5 belongs to P. planiceps ( and ). The nucleotide compositions of the entire sequenced region of Cytb (1100 bp) are A 28.5%, T/U 27.1%, C 31.1%, and G 13.4%.
Figure 2. Cytochrome b based ML tree topology (A) and NJ tree topology (B) of species and subspecies of Prionailurus. The evolutionary distances were computed using the Kimura two-parameter method and are in the units of the number of base substitutions per site. The analysis involved seven nucleotide sequences. Median-joining network (C) by using cytochrome b gene of genus Prionailurus indicating six haplotypes of species and subspecies of Prionailurus. Haplotype 7 belongs to outgroup.
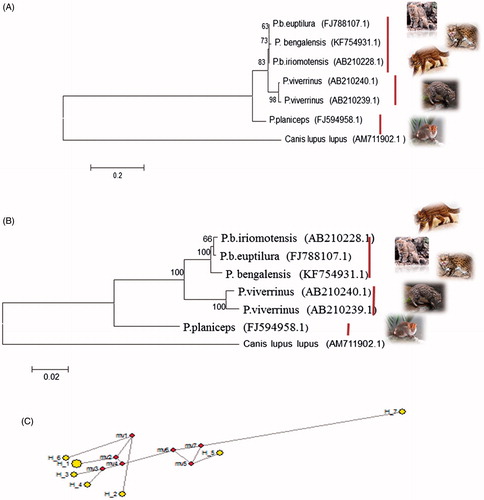
Table 2. Observed DNA diversity in 12Sr RNA and cytochrome b gene.
Phylogenetic study by using maximum likelihood and neighbour joining algorithms (MEGA 6) produced congruent trees () with three clades where all subspecies sequences of P. bengalensis, i.e. irimotensis, euptilurus, and bengalensis form one clade, whereas P. viverrinus and P. planiceps were present in clade B and clade C, respectively, with 100% bootstrap value.
Among the species and subspecies examined in the present study, it was noted that the iriomote cat is a subspecies of P. bengalensis and not a distinct species based on genetic and evolutionary analysis and P.b. iriomotensis and P.b. euptilurus, i.e. Amur leopard cat, shared similar haplotype of 12SrRNA, but P.b. horsfieldi has distinct haplotype for 12SrRNA gene.
Amur Leopard Cat P.b. euptilurus was earlier proposed as a distinct species based on morphological differences from southeast Asian specimens, but Chinese specimens have been shown to be similar to those from southeast Asia (Wozencraft Citation2005). In a molecular study done by Masuda and Yoshida (Citation1995), a clear distinction was noted between northern populations from Tsushima, Korea, Siberia, China and Taiwan and south east Asian populations. If these genetic differences indicate a specific distinction, P.b. euptilurus may yet be a valid species. The Iriomote cat (P.b. iriomotensis) from Japan's Iriomote island was also originally described as a distinct species based on morphology (Imaizumi Citation1967), but based on mt DNA analysis, it is now considered a subspecies of Leopard Cat (Masuda and Yoshida Citation1995; Johnson et al. Citation1999; Wozencraft Citation2005). The present study also reveals that the Iriomote cat is a subspecies of P. bengalensis and genetically similar to P.b. euptilurus. It has been classified as Critically Endangered by IUCN since 2008, as the population size is fewer than 250 thus there is need to apply conservation measures. Recent genetic analysis (Luo et al. Citation2014) suggests species-level distinction between the Indochinese and Sundaic populations of the Leopard Cat. DNA sequence, 12SrRNA, and cytochrome b comparisons were used by Janczewski et al. (Citation1995) to infer phylogenetic relationships among 17 Felid species and supports the use of the two genes for genetically differentiating the species and subspecies for small cats. According to Janczewski et al. (Citation1995), cytochrome b sequences appear to accumulate differences rapidly and then stabilize, whereas 12SrRNA differences with lower number of changes per variable sites seem to accumulate more gradually but steadily.
Conclusions
Cytochrome b is potentially good to determine the genetic difference between the two subspecies of P. bengalensis, i.e. iriomotensis and euptilurus and species of Prionailurus hence in providing the strong scientific proof for identification of the subspecies. However, 12SrRNA is effective to differentiate P. bengalensis horsfieldi from P.b. iriomotensis and P.b. euptilurus.
Acknowledgements
The author is grateful to Dr. Kailash Chandra, Director, Zoological Survey of India for providing facility to conduct the study. The author is also thankful to the in-charge and staff of mammal section, HQ, ZSI Kolkata for providing taxidermy samples for the study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bartlett SE, Davidson WS. 1992. FINS (forensically informative nucleotide sequencing): a procedure for identifying the animal origin of biological specimens. Biotechniques. 12:408-411.
- Barros NDM, Morgante JS. 2007. A simple protocol for the extraction and sequence analysis of DNA from study skin of museum collections. Genet Mol Biol. 30:1181–1185.
- Duckworth JW, Shepherd CR, Semiadi G, Schauenberg P, Sanderson S, Roberton SI, O’Brien TG, Maddox T, Linkie M, Holden J, et al. 2009. Does the fishing cat Prionailurus viverrinus inhabit Sumatra? Cat News. 51:4–9.
- Hall TA. 1990. BioEDIT A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Imaizumi Y. 1967. A new genus and species of cat from Iriomote, Ryukyu Islands. J Mammal Soc Japan. 3:74.
- Janczewski DN, Modi WS, Stephen JC, O’Brien SJ. 1995. Molecular evolution of mitochondrial 12SrRNA and Cytochrome b sequences in the Pantherine lineage of Felidae. Mol Biol Evol. 12:690–707.
- Johnson WE, Ashiki FS, Menotti RM, Driscoll C, Leh C, Sunquist M, Johnston L, Bush M, Wildt D, Yuhki N, et al. 1999. Molecular genetic characterization of two insular Asian cat species, Bornean Bay cat and Iriomote cat. Evolutionary theory and process: modern perspectives. Papers in Honour of Evivatar Nevo, 223. Kluwer Academic Publisher.
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanco FX, Wilson AC. 1989. Dynamics of mitochondrial evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci. 86:06196–06200.
- Luo SJ, Zhang Y, Johnson WE, Miao L, Martelli P, Antunes A, Smith JLD, O’Brien SJ. 2014. Sympatric Asian felid phylogeography reveals a major Indochinese–Sundaic divergence. Mol Ecol. 23:2072–2092.
- Masuda R, Yoshida MC. 1995. Two Japanese wildcats, the Tsushima cat and the Iriomote cat, show the same mitochondrial DNA lineage as the leopard cat Felis bengalensis. Zool Sci. 12:655–659.
- Nowell K, Jackson P. 1996. Fishing cat Prionailurus viverrinus. In: Wild cats: status survey and conservation action plan. Gland, Switzerland: IUCN/SSC Cat Specialist Group.
- Pevsner J. 2009. Bioinformatics and functional genomics online. ISBN:9780470451496.
- Ross J, Hearn AJ, Bernard H, Secoy K, Macdonald D. 2010. A framework for a wild cat action plan for Sabah. Oxford: Global Canopy Programme.
- Sunquist M, Sunquist F. 2002. Wild cats of the world. Chicago: University of Chicago Press; p. 225–232. ISBN 0-226-77999-8.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA 5: molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTRAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 22:4673–4680.
- Vella C, Shelton LM, Gonagle JJ, Stanglein TW. 2002. Robinson's, genetics for cat breeders and veterinarians. 4th ed. Oxford: Butterworth-Heinemann Ltd. ISBN 0-7506-4069-3.
- Werdelin L, Olsson L. 2008. How the leopard got its spots: a phylogenetic view of the evolution of felid coat patterns. Biol J Linn Soc. 62:383–400.
- Wozencraft WC. 2005. Felidae. In: Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. 3rd ed. Baltimore (MD): Johns Hopkins University Press; p. 532–548.
