Abstract
Sanghuang is a polypore mushroom, which has been widely used in oriental medicine. Since recent molecular phylogenetic studies elucidated its species delimitation, Sanghaungporus sanghuang became the official name of this fungus. In this study, the complete sequence of the mitochondrial DNA of S. sanghuang was determined. The whole genome was 112,060 bp containing 14 proteins, 2 ribosomal RNA subunits, and 45 transfer RNAs. The overall GC content of the genome was 23.21%. A neighbour-joining tree based on atp6 sequence data showed its close relationship with the species of Ganoderma and Trametes.
Sanghuang is a polypore belonging to Hymenochaetaceae in Basidiomycota. Its medicinal benefits such as enhancing detoxication and immune boosting have been intensively studied for the past decades (Chen et al. Citation2006; Guo et al. Citation2007; Zhu et al. Citation2008). The term sanghuang has been widely applied to Phellinus linteus (Berk and M.A. Curtis) Teng and its allies in Asian countries (Wu et al. Citation2012). However, detailed morphological (Dai and Xu Citation1998; Lim et al. Citation2003; Dai Citation2010) and molecular phylogenetic studies (Wagner and Fischer Citation2001; Larsson et al. Citation2006; Wu et al. Citation2012; Zhou et al. Citation2016) showed that the correct name is not P. linteus, but Sanghuangporus sanghuang (Sheng H. Wu, T. Hatt. and Y.C. Dai).
The purpose of this study is to characterize the complete mitochondrial genome of S. sanghuang. It will contribute to construct a more comprehensive and natural taxonomic system of this group, as well as to design useful molecular markers to discriminate closely related taxa, which are morphologically similar.
KACC54185 was obtained from the Korean Agricultural Culture Collection (KACC), Republic of Korea. Its mycelium was harvested after 2 weeks incubation on malt extract agar (Difco Laboratories, Detroit, MI, USA). Genomic DNA was extracted using the methodology of Lee and Taylor (Citation1990). Next-generation sequencing was performed using PacBio-RS-II and Hiseq2500 following the manufacturers’ instructions. At first, hierarchical genome assembly process (HGAP) was conducted with the PacBio reads. After mapping the Hiseq reads, de novo assembly of the mitochondrial genomes was completed. GeSeq (Annotation of Organellar Genomes) program (Tillich et al. Citation2017) predicted the mitochondrial gene regions, which were double-checked using blast search and Artemis (Rutherford et al. Citation2000). The complete and annotated mitochondrial genome has been deposited to GenBank with the accession number of MG149786.
The mitochondrial genome of S. sanghuang was represented by linear DNA molecules terminating with inverted repeats (IRs), which may play a role of telomere (Kolesnikov and Gerasimov Citation2012). Linear mitochondrial genomes induced by IRs have been reported in some fungal species (Forget et al. Citation2002; Salavirta et al. Citation2014). The total length was 112,060 bp, which is the second largest among the hitherto reported mitogenomes of polypores (Wang et al. Citation2016). It contains 2 ribosomal RNA subunits (rns and rnl), 45 transfer RNA genes, and 14 protein-coding genes of an apocytochrome b (cob), three cytochrome c oxidases (cox1, cox2, and cox3), three ATPases (atp6, atp8, and atp9), and seven NADH dehydrogenases (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6). The base composition was 38.37% A, 38.41% T, 12.14%G, and 11.07%C with an AT bias of 76.78%.
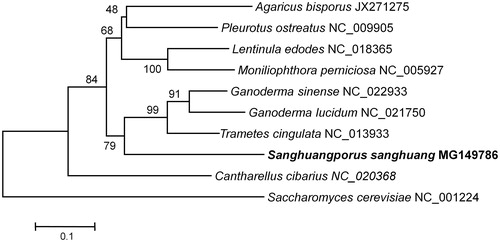
To ascertain the position of S. sanghuang within the Agaricomycotina, a neighbour-joining tree was constructed with MEGA6 (Tamura et al. Citation2013) using atp6 of representative species belonging to Agaricomycotina. The inferred tree represented that sanghuang (Hymenochaetaceae) forms a sister clade of Ganoderma lucidum, G. sinense (Ganodermataceae), and Trametes cingulata (Polyporaceae) (). This is consistent with the traditional classification of the Agaricomycotina with robust bootstrap supports.
Acknowledgements
The authors would like to appreciate Dr. Seung-Beom Hong of the Korean Agricultural Culture Collection, Rural Development Administration for his assistance in access to the Sanghuangporus sanghaung stain KACC54185.
Disclosure statement
The authors have declared that no competing interests exist.
Additional information
Funding
References
- Chen W, He FY, Li YQ. 2006. The apoptosis effect of hispolon from Phellinus linteus (Berkeley & Curtis) Teng on human epidermoid KB cells. J Ethnopharmacol. 105:280–285.
- Dai YC. 2010. Hymenochaetaceae (Basidiomycota) in China. Fungal Diversity. 45:131–343.
- Dai YC, Xu MQ. 1998. Studies on the medicinal polypore, Phellinus baumii, and its kin, P. linteus. Mycotaxon. 67:191–200.
- Forget L, Ustinova J, Wang Z, Huss VA, Lang BF. 2002. Hyaloraphidium curvatum: a linear mitochondrial genome, tRNA editing, and an evolutionary link to lower fungi. Mol Biol Evol. 19:310–319.
- Guo J, Zhu T, Collins L, Xiao ZX, Kim SH, Chen CY. 2007. Modulation of lung cancer growth arrest and apoptosis by Phellinus linteus. Mol Carcinog. 46:144–154.
- Kolesnikov AA, Gerasimov ES. 2012. Diversity of mitochondrial genome organization. Biochemistry Mosc. 77:1424.
- Larsson KH, Parmasto E, Fischer M, Langer E, Nakasone KK, Redhead SA. 2006. Hymenochaetales: a molecular phylogeny for the hymenochaetoid clade. Mycologia. 98:926–936.
- Lee S, Taylor J. 1990. Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: a guide to methods and applications. San Diego, USA: Academic Press; p. 315–322.
- Lim YW, Lee JS, Jung HS. 2003. Type studies on Phellinus baumii and Phellinus linteus. Mycotaxon. 85:201–210.
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics. 16:944–945.
- Salavirta H, Oksanen I, Kuuskeri J, Mäkelä M, Laine P, Paulin L, Lundell T. 2014. Mitochondrial Genome of Phlebia radiata is the second largest (156 kbp) among fungi and features signs of genome flexibility and recent recombination events. PLos One. 9:e97141.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucl Acid Res. 45:W6–W11.
- Wagner T, Fischer M. 2001. Natural groups and a revised system for the European poroid Hymenochaetales (Basidiomycota) supported by nLSU rDNA sequence data. Mycol Res. 105:773–782.
- Wang XC, Shao J, Liu C. 2016. The complete mitochondrial genome of the medicinal fungus Ganoderma applanatum (Polyporales, Basidiomycota). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2813–2814.
- Wu SH, Dai YC, Hattori T, Yu TW, Wang DM, Parmasto E, Chang HY, Shih SY. 2012. Species clarification for the medicinally valuable ‘sanghuang’ mushroom. Bot Stud. 53:135–149.
- Zhou LW, Vlasak J, Decock C, Assefa A, Stenlid J, Abate D, Wu SH, Dai YC. 2016. Global diversity and taxonomy of the Inonotus linteus complex (Hymenochaetales, Basidiomycota): Sanghuangporus gen. nov., Tropicoporus excentrodendri and T. guanacastensis gen. et spp. nov., and 17 new combinations. Fungal Diversity. 77:335–347.
- Zhu T, Kim SH, Chen CY. 2008. A medicinal mushroom: Phellinus linteus. Curr Med Chem. 15:1330–1335.