Abstract
The wax-leafed Viburnum japonicum (Adoxaceae) is an evergreen shrub distributed in Japan, Korea, and Taiwan. We sequenced its complete chloroplast (cp) genome to examine its phylogenetic relationship within Dipsacales. This genome is 158,614 bp long and features a large single-copy region (87,059 bp) and a small single-copy region (18,523 bp), separated by two inverted-repeat regions (26,516 bp each). It contains 128 genes, including 84 coding genes, eight rRNAs, and 36 tRNAs. The overall GC content is 38.1%. Our phylogenetic tree showed that V. japonicum is closely related to V. utile and is clustered together with four species in the family Adoxaceae.
Within Dipsacales, the Viburnum genus has traditionally been regarded as a group within Caprifoliaceae along with Sambucus and Sinadoxa (Cronquist Citation1988). The latter two also belong to Adoxaceae, under the classification of APG (Donoghue et al. Citation2001). Analyses of molecular and morphological data suggest that Viburnum is the basal-most lineage in the family Adoxaceae (Jacobs et al. Citation2010). Therefore, understanding the evolutionary relationships of Viburnum, a major basal group, would provide a powerful phylogenetic utility and, in particular, convincing evidence for early-diverging Adoxaceae. Viburnum japonicum (Thunb.) Spreng. is an evergreen shrub distributed in the subtropical regions of Korea, Japan, and Taiwan (Yang and Chiu Citation1998; Hong and Im Citation2003). Here, we report its complete chloroplast (cp) genome sequence.
We collected samples of V. japonicum from Is. Gageo-do, Jeollanam-do Province, Korea (N 34° 03′ 59″, E 125° 07′ 04″). The voucher specimen was stored at the herbarium in the Department of Biology Education, Chonnam National University (BEC: Lee. 16101504). Total genomic DNA was extracted with a DNeasy Plant Mini Kit (Qiagen, Seoul, Korea), and pair-end sequencing (300 × 300 bp) was performed using the Illumina Miseq platform (LAS, Seoul, Korea). This presented 13,920,280 raw reads. After trimming the sequences, clean reads were mapped with the reference cp genome for V. utile (NC_032296), using Geneious 10.2.3 (Kearse et al. Citation2012). On that genome, 93,629 reads were assembled with an average of 177X coverage (max: 286X, min: 60X). The annotation was separately performed using DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005). The annotated cp genome sequence was then deposited in GenBank with Accession Number MH036493. To construct the phylogenetic tree, we performed maximum likelihood (ML) analysis with RAxML v.8.0 (Stamatakis Citation2014), using complete cpDNAs of nine related species from the NCBI database aligned with MAFFT (Katoh and Toh Citation2010).
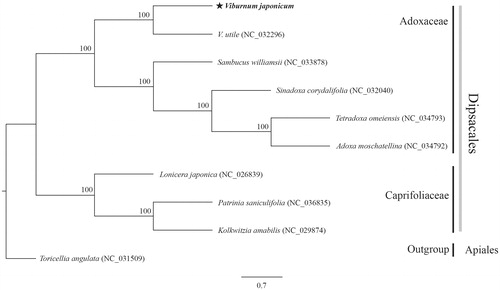
The cp genome of V. japonicum is 158,614 bp long, with 38.1% GC content. It has two inverted-repeat (IR) regions of 26,516 bp that separate a large single-copy region (87,059 bp) and a small single-copy region (18,523 bp). The genome contains 128 genes: 84 protein-coding genes, eight rRNA genes, and 36 tRNA genes. Seventeen genes are duplicated in the IR region. The ML phylogenetic tree, with 1000 bootstrap replications, is monophyletic with two independent clades in Dipsacales. Viburnum japonicum is closely related to V. utile and clusters together with four species in the family Adoxaceae (). The chloroplast genome of V. japonicum provides information for further phylogenetic studies and genome resources within Dipsacales.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Cronquist A. 1988. The evolution and classification of flowering plants. 2nd ed. New York: New York Botanical Garden
- Donoghue MJ, Eriksson T, Reeves PA, Richard GO. 2001. Phylogeny and phylogenetic taxonomy of Dipsacales, with special reference to Sinadoxa and Tetradoxa (Adoxaceae). Harv Pap Bot. 6:459–479.
- Hong HW, Im HT. 2003. Viburnum japonicum (Caprifoliaceae): an unrecorded species in Korea. Korean J Pl Taxon. 33:271–277. (in Korean).
- Jacobs B, Huysmans S, Smets E. 2010. Evolution and systematic value of fruit and seed characters in Adoxaceae (Dipsacales). Taxon. 59:850–866.
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 26:1899–1900.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28: 1647–1649.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yang KC, Chiu ST, 1998. Caprifoliaceae. In: Huang TC, et al., editors. Flora of Taiwan. 2nd ed. Vol. 4. Taipei: Editorial Committee of the Flora of Taiwan. p. 738–759.