Abstract
Jilin ginseng, Panax ginseng C. A. Meyer, belongs to the Araliaceae family. It is known as the number one medicinal herb and one of the three native treasures in Northern China and has been cultivated in China for over 2000 years. Jilin ginseng is the staple ginseng in China, which contributes to 85% of the ginseng production in the country, thus becoming one of the most important, rapidly increasing industries in both the Province of Jilin and China. In this study, the complete chloroplast genome of Panax ginseng C. A. Meyer was determined by the next generation sequencing. The complete chloroplast genome of Jilin ginseng (P. ginseng C. A. Meyer) was 156,286 bp in length and displays a typical quadripartite structure of the large (LSC, 87,127 bp) and small (SSC, 18,329 bp) single-copy regions, separated by a pair of inverted repeat regions (IRs, 25,415 bp each). It harbors 132 functional genes, including 132 protein-coding genes, 37 transfer RNA, and 8 ribosomal RNA genes species. The overall nucleotide composition was: 30.6% A, 31.3% T, 19.4% C, and 18.7% G, with a total G + C content of 38.1%. Phylogenetic relationship analysis shows that P. ginseng closely related to Panax quinquefolius L.
Jilin ginseng, Panax ginseng C. A. Meyer, traditionally known in East Asia, particularly in China, Korea, and Japan, as a medicinal herb (Yun Citation2001). It is a perennial of the Araliaceae family and has been cultivated in China for over 2000 years. Chinese ginseng is mainly grown in Jilin Province, China, and known as Jilin ginseng, and estimated to produce 85% and 70% of the ginseng of China and the world. Modern pharmacological research has focused on the ginsenosides, the major bioactive compound for human health, such as recovery and promotion of vitality, improvement of immune and metabolism systems, regulation of central nervous system, etc (Lee et al. Citation2008; Su et al. Citation2018). In this study, we used the next generation sequencing to get the complete chloroplast genome of ginseng, in order to provide information for the study of the origin of medicinal plants in evolution.
The specimen of P. ginseng was isolated from Jilin Agricultural University (Jilin ginseng test field in Changchun, Jilin, China) (125.40E; 43.82N) and the DNA of P. ginseng was stored in Jilin Agricultural University College of Life Science (No. JLAUCLS5). The Jilin ginseng DNA sample was sequenced using next generation sequencing (Illumina Hiseq4000, CA, USA). The P. ginseng complete chloroplast genome was preliminarily annotated using the DOGMA (Dual Organellar GenoMe Annotator) online program (Wyman et al. Citation2004), with default settings to identify protein-coding genes, rRNAs and tRNAs based on the Plant Plastid Code and BLAST homology searches. The secondary structures of transfer RNA (tRNA) genes were identified by using the ARAGORN (Laslett and Canback Citation2004) or through manually visual inspection.
The chloroplast genome sequence of P. ginseng is a closed-circular molecule of 156,286 bp in length, which is almost the same as the Panax quinquefoli chloroplast (156,359 bp). It displayed 132 functional genes set, which is observed in this plant chloroplast, including 87 PCGs, 37 tRNA genes (one for each amino acid, two each for Valine, Asparagine, Serine and Threonine, three each for Leucine and Arginine, three each for Methionine), and 8 genes for ribosomal RNA subunits (two each for rrn16, rrn23, rrn4.5, and rrn5). The chloroplast organization of P. ginseng is compact, encoding 26,108 bp functional regions (including D-loop region). The annotated chloroplast genome was submitted to GenBank database under accession No.MH049735.
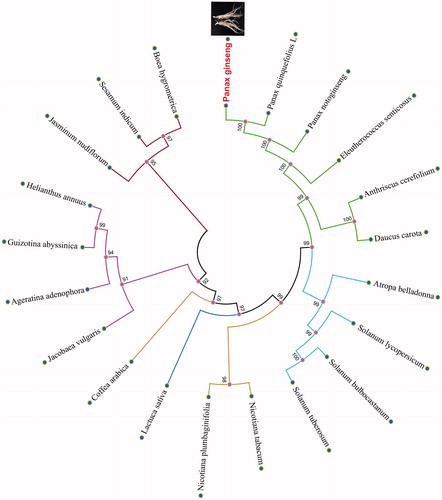
We selected other 20 related complete chloroplast genomes from GenBank to assess the phylogenetic relationship between them. The genome-wide alignment of all plant complete chloroplast genomes was done by HomBlocks (Bi et al. Citation2017). The phylogenetic trees were reconstructed using maximum likelihood (ML) and neighbour-joining (NJ) methods. ML analysis were performed using RaxML-8.2.4 (Stamatakis Citation2014), of which the bootstrap values were calculated using 1000 replicates to assess node support. NJ phylogenetic tree was constructed using MEGA7 with 1000 bootstrap replicate (Kumar et al. Citation2016). All the nodes were inferred with strong support by the ML and NJ methods. As shown in the phylogenetic tree (), P. ginseng showed the closest with P. quinquefolius L.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2017. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 1:110.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16.
- Lee ST, Chu K, Sim JY, Heo JH, Kim M. 2008. Panax ginseng enhances cognitive performance in Alzheimer disease. Alzheimer Dis Assoc Disord. 22:222–226.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Su JE, Kim KT, Paik HD. 2018. Microbial bioconversion of ginsenosides in Panax ginseng and their improved bioactivities. Food Rev Int. 1–15.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yun TK. 2001. Brief introduction of Panax ginseng C.A. Meyer. J Korean Med Sci. 16 (Suppl):S3–S5.