Abstract
The complete mitochondrial genome of the oyster Crassostrea belcheri from the Cần Giò’ mangrove in Vietnam has been sequenced. It consists of a circular DNA molecule of 21020 base pairs (bp), coding for 12 proteins, 20 transfer RNAs, and two ribosomal RNAs. Like the mitogenomes of Crassostrea iredalei and Crassostrea sp. DB1, it contains a non-coding region and two ORFs. The C. belcheri mitogenome provides information that could improve the molecular phylogeny of Asian oysters and be useful to the development of oyster aquaculture in South East Asia.
The tropical oyster Crassostrea belcheri is cultured commercially in South-East Asia (Tan and Wong Citation1996; Klinbuga et al. Citation2000, Citation2003, Citation2005) and Indonesian archipelago (Yoo and Ryu Citation1984). Its commercialization is an important activity in the Cần Giò’ mangrove, located near Hồ Chí Minh City in Vietnam, where it is sold under the vernacular name of Hàu Dép. In this study, we present the complete mitogenome of C. belcheri. The specimen was collected in the mangrove in September 2017, in the Lò Vôi river (10°25′55″N; 106°54′530″E), the remaining flesh and shells being conserved at Saigon University. DNA was extracted following a chloroform-isopropanol protocol, and sequencing was provided by the Beijing Genomic Institute, consisting of 4 Gb of 150-bp paired-end reads from fragments of 300 bp, obtained on an Illumina HiSeq 4000. Data were assembled using Ray 2.3.1 (Boisvert et al. Citation2010) with a k-mer of 31.
The C. belcheri mitogenome is 21020 bp long (GenBank accession no. MH051332), making it the third longest mitogenome sequenced among the genus Crassostrea (Wu et al. Citation2010, Citation2012). The nucleotide composition of this mitogenome is A 28.3%, C 14.3%, G 21.4%, T 36%. It encodes 12 proteins, 20 transfer RNAs, and two ribosomal RNAs. The rrnL gene is split into two parts and the rrnS gene is duplicated, both features being commonly found among Asian oysters (Wu et al. Citation2010). As in other Crassostrea mitogenomes, there is a major non-coding region whose size is 797 bp in C. belcheri. Also, the C. belcheri mitogenome shares with Crassostrea iredalei (FJ841967) and Crassostrea sp. DB1 (JQ060958) two ORFs originating from the duplication of the NAD2 gene. The latter ORFs are separated from one another by a putative suppressor tRNA gene.
A maximum-likelihood phylogenetic analysis was performed using the complete mitogenome sequences of 11 Crassostrea species and the resulting tree was rooted using the mitogenome of Ostrea edulis (). C. belcheri was recovered between a strongly supported cluster containing C. hongkongensis, C. ariakensis, C. nippona, C. sikamea, C. angulate, and C. gigas, which all have mitogenomes very similar in size and nucleotide composition, and a second robust cluster containing C. iredalei and Crassostrea sp. DB1, which feature the longest mitogenomes and the two conserved ORFs. The highly supported clade formed by all nine Crassostrea species was sister to that containing C. virginica and C. gasar, both from the American continent.
Figure 1. Maximum likelihood phylogeny using the complete mitogenomes of Crassostrea belcheri and other species of oysters, with Ostrea edulis as an outgroup. The tree with the highest log likelihood (−161490.9077) is shown. Numbers next to nodes are support values obtained after 1000 bootstrap replicates.
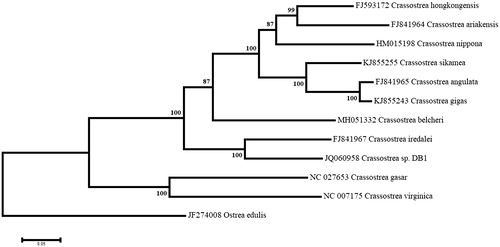
The C. belcheri mitogenome thus provides new information regarding the phylogeny and genome evolution of Asian Crassostrea species. It may also serve as a reliable reference sequence for accurate molecular barcoding of the different populations of C. belcheri and identification of specimens, a tool that may be helpful for the development of the aquaculture of this species in South East Asia.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Boisvert S, Laviolette F, Corbeil J. 2010. Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol. 17:1519–1533.
- Klinbuga S, Ampayup P, Tassanakajon A, Jarayabhand P, Yoosukh W. 2000. Development of species-specific markers of the tropical oyster (Crassostrea belcheri) in Thailand. Mar Biotechnol. 2:476–484.
- Klinbuga S, Khamnantong N, Puanglarp N, Jarayabhand P, Yoosukh W, Menasveta P. 2005. Molecular Taxonomy of Cupped Oysters (Crassostrea, Saccostrea, and Striostrea) in Thailand Based on COI, 16S, and 18S rDNA Polymorphism. Mar Biotechnol. 7:306–317.
- Klinbuga S, Khamnantong N, Tassanakajon A, Puanglarp N, Jarayabhand P, Yoosukh W. 2003. Molecular Genetic Identification Tools for Three Commercially Cultured Oysters (Crassostrea belcheri, Crassostrea iredalei, and Saccostrea cucullata) in Thailand. Mar Biotechnol. 5:27–36.
- Tan SH, Wong TM. 1996. Effect of salinity on hatching, larval growth, survival and settling in the tropical oyster Crassostrea belcheri (Sowerby). Aquaculture. 145:129–139.
- Wu X, Li X, Li L, Xu X, Xia X, Yu Z. 2012. New features of Asian Crassostrea oyster mitochondrial genomes: a novel alloacceptor tRNA gene recruitment ant two novels ORFs. Gene. 507:112–118.
- Wu X, Xu X, Yu Z, Wei Z, Xia J. 2010. Comparison of seven Crassostrea mitogenomes and phylogenetic analyses. Mol Phylogenet Evol. 57:448–454.
- Yoo SK, Ryu HY. 1984. Comparative morphological characteristics of mangrove oysters. Bull Korean Fish Soc. 17:321–326.
