Abstract
Half-fin anchovy (Setipinna tenuifilis) is one of the most important economic fishes around the world. In the present study, we determined the complete mitochondrial DNA sequence and organization of S. tenuifilis. The entire mitochondrial genome is a circular-molecule of 16,215 bp in length, which encodes 37 genes in all. These genes comprise 13 protein-coding genes (ATP6 and 8, COI–III, Cytb, ND1-6, and 4L), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (12S and 16S rRNAs), with gene arrangement and content basically identical to those of other species of Engraulidae. The result of phylogenetic analysis strongly supported that S. tenuifilis was first clustered together with Setipinna melanochir and formed a monophyly in the genus Coilia, and then they constituted a sister-group relationship with two genus Engraulis, and Stolephorus. It concluded that the S. tenuifilis should be classified into the genus Setipinna. The present study also revealed the phylogenetic relationship of this genus at molecular levels. The complete mitochondrial genome sequence of S. tenuifilis can provide basic information for the studies on molecular taxonomy and phylogeny of teleost fishes.
Half-fin anchovy (Setipinna tenuifilis), one of the most important economic fishes, is a highly migratory species distributed in the Indian Ocean and Western Pacific Ocean. Till now, we know little about mitogenome of S. tenuifilis. Thus, we determined the complete mitogenome sequence and organization of S. tenuifilis here.
Setipinna tenuifilis specimens were collected from Shanghai, China and deposited in East China Sea Fisheries Research Institute. The complete mitogenome sequence of S. tenuifilis was determined by polymerase chain reaction amplification and sequencing, using 13 pairs of primers which were designed based on mitogenome sequence of Setipinna melanochir (GenBank number AP011565.1), a closely related species of S. tenuifilis. The complete mitogenome of S. tenuifilis was deposited in GenBank database with accession number MH037012. Muscle tissues were persevered in 95% ethanol for DNA extraction. Total genomic DNA was extracted from muscle tissue by standard phenol–chloroform procedure (Sambrook et al. Citation1989).
The complete mitogenome of S. tenuifilis was determined to be 16,215 bp, comprising 37 coding and two non-coding regions. The 37 coding regions include 13 protein-coding genes (ATP6 and 8, COI–III, Cytb, ND1-6, and 4L), 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (12S and 16S rRNAs), and two non-coding regions consist of light-strand replication origin (OL) and control region (CR). Except for one protein-coding gene ND6 and eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro), all other genes are encoded on the heavy strand, which are in accordance with other teleost mitogenomes (Zhu et al. Citation2013).
The overall nucleotide base composition of S. tenuifilis mitogenome is as follows: A, 30.90%; G, 15.70%; T, 25.00%; and C, 28.40%, respectively, showing an obvious anti-G bias as appeared in other teleost species (Jondeung et al. Citation2007).
The CR of S. tenuifilis is 1254 bp in length, with tRNAPro and tRNAPhe at its two ends. The OL is 29 bp long, locating in a cluster of the tRNATrp-tRNAAla-tRNAAsn-tRNACys-tRNATyr region (WANCY region, ).
Table 1. Characteristics of the mitochondrial genome of Setipinna tenuifilis.
All protein-coding genes in S. tenuifilis have a methionine (ATG) start codon except for COI and ND 6, which starts with ATC and AAC. The 13 protein-coding genes (ND1, ND2, COI, COII, ATP8, ATP6, COIII, ND3, ND4L, ND4, ND5, ND6, and Cytb) are ended with CTA, ATT, ATG, CAA, CAT, CTG, ACT, AAG, CGT, CGG, ATC, TAC, and TTC ().
There are some overlaps existing in S. tenuifilis mitogenome. For 13 protein-coding genes, there are two overlaps detected in ATP8-ATP6 and ND4-ND4L. Among 22 tRNAs, three overlaps are found in tRNAIle-tRNAGln, tRNAGln-tRNAMet, and tRNAThr-tRNAPro ().
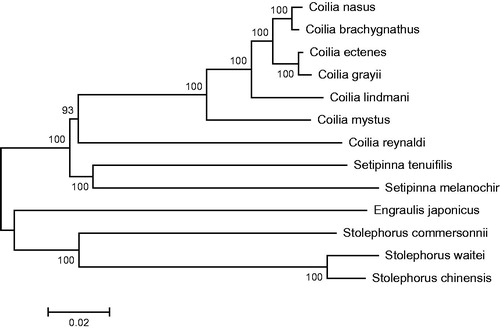
Phylogenetic analysis was performed by MEGA 6.06 (Tamura et al. Citation2013) based on complete mitogenome sequence of S. tenuifilis and those of 13 closely related species belonging to four genus Engraulis, Stolephorus, Setipinna, and Coilia. The neighbour-joining tree () showed that S. tenuifilis first clustered together with Setipinna melanochir and formed a monophyly in the genus Coilia, and then they constituted a sister-group relationship with other two genus. This result strongly supported that S. tenuifilis should be classified into the genus Setipinna. This study also revealed the phylogenetic relationship of the genus Coilia at molecular levels.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Jondeung A, Sangthong P, Zardoya R. 2007. The complete mitochondrial DNA sequence of the Mekong giant catfish (Pangasianodon gigas), and the phylogenetic relationships among Siluriformes. Gene. 387:49–57.
- Sambrook J, Maniatis TE, Fritsch EF. 1989. Molecular cloning: a laboratory manual. Vol. 1. New York: Cold Spring Harbor Laboratory Press.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. Mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhu YX, Chen Y, Cheng QQ, Qiao HY, Chen WM. 2013. The complete mitochondrial genome sequence of Schizothorax macropogon (Cypriniformes: Cyprinidae). Mitochondrial DNA. 24:237–239.