Abstract
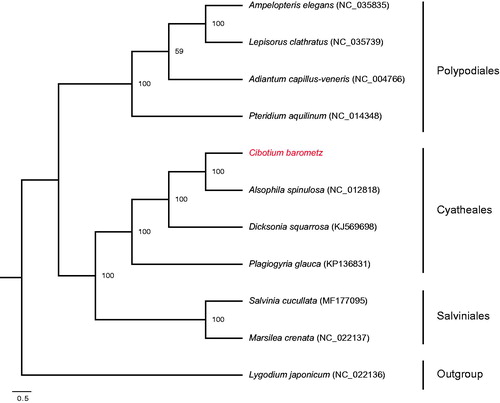
The complete chloroplast (cp) genome sequence of Cibotium barometz, an endangered CITES medicinal fern, was determined by Illumina sequencing. Its genome is a circular molecule of 166,027 bp in length, including a pair of inverted repeats (IRs) of 29,158 bp, a large single-copy (LSC) region of 85,665 bp, and a small single-copy (SSC) region of 22,046 bp. The genome encodes 118 unique genes, involving 84 protein-coding genes, 28 tRNA genes, four rRNA genes, and two pseudogenes. Maximum likelihood tree showed that C. barometz was closely related to Alsophila spinulosa. This investigation lays solid foundations for endangered medicinal resource conservation and fern phylogenetic studies.
Cibotium barometz (L.) J. Sm is a large terrestrial tree fern with fronds up to 2 metres tall in Cibotiaceae (Smith et al. Citation2006), previously in Dicksoniaceae. As an indicator species of acidic soils, it is widely distributed in south subtropical and tropical regions, such as Guangdong, Guangxi, Guizhou, Sichuan, and Yunnan in China (Zhang and Nishida Citation2013). This fern prefers warm humid environments at elevations up to 1600 m (Zhang and Nishida Citation2013). Its most prominent feature reflects in stump-like rhizome covered by long, soft, golden-yellow hairs, showing like a golden-haired dog. The species is not only an ornamental fern, but also a famous traditional Chinese herbal plant known as ‘Gouji’ with anti-inflammation and anti-osteoporosis activities (Cuong et al. Citation2009; Wu and Yang Citation2009; Zhao et al. Citation2011). Due to uncontrolled collection, C. barometz is listed as an endangered plant of National Protection Grade II, and in Appendix II of the Convention on International Trade in Endangered Species of Wild Fauna and Flora as well (CITES Citation2017). In addition, Cibotium is subjected to much confusion and revision (Zhang and Nishida Citation2013). Hence, the acquirement of whole chloroplast (cp) genome of C. barometz will contribute to resource protection and phylogenetics.
The plant sample was harvested from South China Botanical Garden, Chinese Academy of Sciences. Voucher specimen is held by the Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 20161011). Genomic DNA was extracted from fresh leaves using Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China). Approximately 300–500 bp paired-end library was constructed and sequenced in Hiseq 2500 platform (Illumina Inc., San Diego, CA, USA). After adapters or low-quality reads were removed by Trimmomatic (Bolger et al., Citation2014), a genome was obtained through de novo assembly using Velvet v1.2.07 (Zerbino and Birney Citation2008). DOGMA (Wyman et al. Citation2004) and tRNAscan-SE 1.21 (Schattner et al. Citation2005) were applied to perform annotation. Maximum-likelihood analysis including 11 ferns was conducted based on complete cp genome sequences using RAxMLv.8.0 (Stamatakis Citation2014) with 1000 bootstrap replicates. Lygodium japonicum was selected as outgroup.
The complete chloroplast genome of C. barometz is 166,027 bp in length, with a pair of IR regions of 29,158 bp that separate an LSC region of 85,665 bp and an SSC region of 22,046 bp (GenBank accession number: MH105066). It encodes 131 genes, of which 118 genes are unique, including 84 protein-coding genes (PCGs), 28 tRNA genes, four rRNA genes, and two pseudogenes. Thirteen genes are duplicated, containing four rRNA genes (rrn5, rrn4.5, rrn23, and rrn16), five tRNA genes (trnN-GUU, trnH-GUG, trnI-GAU, trnA-UGC, and trnR-ACG), three PCGs (psbA, rps7, and rps12), and one pseudogenes (ycf2). Nine PCGs and five tRNA genes have one intron, whereas two PCGs and one trans-splicing gene contain two introns. The overall GC content is 41.7%. ML tree revealed that C. barometz was sister to Alsophila spinulosa (). The complete chloroplast genome of C. barometz lays solid foundations for endangered medicinal resource conservation and fern phylogenetic studies.
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- CITES. 2017. Convention on International Trade in Endangered Species of Wild Fauna and Flora. Appendices I, II, and III. [accessed 2018 mar 15]. Available from: https://cites.org/eng/app/appendices.php
- Cuong NX, Minh CV, Kiem PV, Huong HT, Ban NK, Nhiem NX, Tung NH, Jung JW, Kim HJ, Kim SY, et al. 2009. Inhibitors of osteoclast formation from rhizomes of Cibotium barometz. J Nat Prod. 72:1673–1677.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Smith AR, Pryer KM, Schuettpelz E, Korall P, Schneider H, Wolf PG. 2006. A classification for extant ferns. Taxon. 55:705–731.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wu Q, Yang XW. 2009. The constituents of Cibotium barometz and their permeability in the human Caco-2 monolayer cell model. J Ethnopharmacol. 125:417–422.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang XC, Nishida H. 2013. Cibotiaceae. In: Wu ZY, Raven PH, Hong DY, eds., Flora of China, Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 132–133.
- Zhao X, Wu ZX, Zhang Y, Yan YB, He Q, Cao PC, Lei W. 2011. Anti-osteoporosis activity of Cibotium barometz extract on ovariectomy-induced bone loss in rats. J Ethnopharmacol. 137:1083–1088.