Abstract
The complete mitogenome of the stalk-forming diatom Didymosphenia geminata collected from Mineral County, WV, USA was sequenced on the Ion Torrent PGM and Proton sequencers. The D. geminata mitogenome is 37,765 bp and encodes 35 protein coding genes, 25 tRNAs, and both large and small subunit ribosomal RNA genes. The nad11 gene is split into two domains as observed in Phaeodactylum tricornutum, and D. geminata also lacks the large repeat region found in the P. tricornutum mitogenome. Gene order and content within the D. geminata mitogenome is similar to the diatom Berkeleya fennica.
The freshwater stalk-forming diatom Didymosphenia geminata has been the focus of intense scientific investigations over the last decade because of the increasing frequency of large scale D. geminata ‘blooms’ in rivers and streams worldwide. The cause of these blooms remains poorly understood, though streams with low inorganic phosphorous and clear and cool water appear to be most at risk (Bergey and Spaulding Citation2015). A paucity of genetic data exists for D. geminata, but is necessary to better understand how D. geminata nuisance blooms form, as well as to better understand patterns of phylogeographic structure. Here, we describe the complete mitogenome of D. geminata to move forward genomic study of this nuisance species.
A 2 cm3 sample of D. geminata mat was obtained from the tailwaters of Jennings Randolph reservoir (39.4318°N –79.1152°W) in Mineral County, WV, March 2014. The sample is being stored at the U.S. Geological Survey Leetown Science Center, Kearneysville, WV, and 300 intact D. geminata detached from their stalks were isolated from this mat sample by mouth pipette for analysis. These cells were incubated in a lysozyme solution (20 mM Tris HCl pH 8, 2 mM sodium EDTA, 20 mg/mL lysozyme) for 10 minutes at 25 °C, and rinsed with 0.25X TE to reduce bacterial contamination in the subsequent Qiagen DNEasy DNA extraction. Libraries were prepared for single end sequencing on both the Ion Torrent PGM and Proton sequencers (Thermo Fisher Scientific, Frederick, MD, USA). De novo assembly of 14,018,492 sequence reads from the PGM and 39,227,730 sequence reads from the Proton in CLC Genomics Workbench (Qiagen, Frederick, MD, USA) resulted in four separate (but overlapping) contigs of mtDNA origin. A consensus of the overlapping contigs was generated and the reads were mapped back to this consensus resulting in a complete circular mapping mitogenome of 37,765 bp with an average coverage of 1218. Annotation of protein coding, ribosomal genes, and tRNAs was accomplished using the program MFANNOT (Beck and Lang Citation2010). A partitioned maximum likelihood phylogenetic tree with 1000 ultrafast bootstrap resamplings (Hoang et al. Citation2018) was created in the program IQ-Tree (Nguyen et al. Citation2015) using a MUSCLE alignment of 6817 amino acids (Edgar Citation2004) generated in the program TranslatorX (Abascal et al. Citation2010) of 27 shared protein coding genes (atp6, atp9, cob, cox1, cox2, cox3, nad11, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, rpl14, rpl2, rpl5, rps10, rps12, rps13, rps14, rps19, rps3, rps4, rps8) from 7 other selected members of the class Bacillariophyceae with complete mitogenomes. The best partitioning scheme for the analysis was determined with PartitionFinder2 (Lanfear et al. Citation2017).
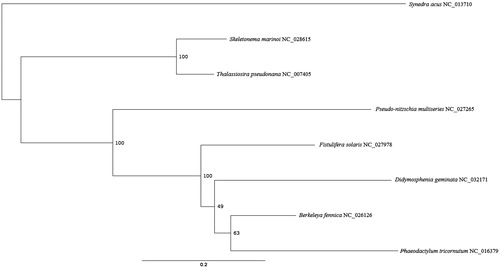
The D. geminata mitogenome (GB KX889125) codes for the small and large subunit rRNAs, 35 protein coding genes (atp6, 8, 9; cob; cox1, 2, 3; nad1-4, 4L, 5-7, 9,11a,11b; rpl2, 5, 6, 14, 16; rps2-4, 7, 8, 10-14, 19; tatC, tatA), and 25 tRNAs. The gene order was nearly identical to the mitogenome of the diatom Berkeleya fennica (An et al. Citation2016), with the exception of D. geminata having tRNA-Gln before cox1, and an extra tRNA-Ile after nad2. Start codons included ATG, GTG, and TTG, while stop codons included TAA, TAG, and TAA. Like within Phaeodactylum tricornutum, the nad11 gene is split into two domains in D. geminata (Oudot-Le Secq and Green Citation2011). However, D. geminata does not possess a large repeat region as observed in the P. tricornutum mitogenome. The maximum likelihood tree indicates that D. geminata forms a monophyletic clade with Fistulifera solaris, Berkeleya fennica, and Phaeodactylum tricornutum, but there is low bootstrap support for the relationships among D. geminata, Berkeleya fennica, and Phaeodactylum tricornutum ().
Figure 1. Partitioned maximum likelihood unrooted phylogenetic tree constructed in IQ-Tree (Nguyen et al. Citation2015) of 6817 amino acid positions with 1000 ultrafast bootstrap resamplings (Hoang et al. Citation2018) indicating the relationship of Didymosphenia geminata with seven other members of the class Bacillariophyceae based on 27 shared protein coding genes from the mitochondrial genome (atp6, atp9, cob, cox1, cox2, cox3, nad11, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, nad9, rpl14, rpl2, rpl5, rps10, rps12, rps13, rps14, rps19, rps3, rps4, rps8). The partitioning scheme with the best corrected AIC score in PartitionFinder2 was used (Lanfear et al. Citation2017). GenBank accession numbers are next to the species names.
Acknowledgements
The authors would like to thank Jim Hedrick of the WV Department of Natural Resources, and US Army Corps of Engineers staff at Jennings Randolph Lake, Mineral County, WV for helping coordinate collection of D. geminata.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:7–13.
- An SM, Noh JH, Choi DH, Lee JH, Yang EC. 2016. Repeat region absent in mitochondrial genome of tube-dwelling diatom Berkeleya fennica (Naviculales, Bacillariophyceae). Mitochondrial DNA A. 27(3):2137–2138.
- Beck N, Lang B. 2010. MFannot, organelle genome annotation webserver. [accessed 2016 June 16]. http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl
- Bergey EA, Spaulding SA. 2015. Didymosphenia: it’s more complicated. Aquatic Invasions. 6:249–262.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35:518–522.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34:772–773.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-Tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Oudot-Le Secq MP, Green BR. 2011. Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene. 476:20–26.
