Abstract
Moschus berezovskii is an endangered species, but its captive populations are valuable on musk secretions in traditional Chinese medicine and perfume manufacture. The mitogenome of M. berezovskii was 16,353 bp in size. Stop codons in 13 PCGs were all typical types except incomplete stop codon T for COX3, ND2 and ND4, and TA for ND3. No tandem repeat was found in control region. Phylogenetic analysis indicated that Moschidae has the closest relationship with Bovidae. We supported that M. berezovskii should be categorized into two subspecies, and suggested that the status of M. chrysogaster JQ608470 should be further investigated.
Forest Musk Deer, Moschus berezovskii was listed as a threatened species in the Red List (IUCN Citation2017) and categorized as a first-degree national protected species in China. Once abundant, poaching and habitat loss had led to a dramatic decrease in wild population numbers (Gang et al. Citation2015) prompting to establish captive breeding populations for musk secretions. Nevertheless, during the long process of captive breeding, historical records about the genetic backgrounds of many captive populations were either lost or incomplete. Therefore, understanding the genetic diversity and phyletic evolution of the captive populations are the two most important tasks for excellent provenance selection (Peng et al. Citation2008).
Sample (voucher no. LS018) of captive M. berezovskii was deposited in the animal specimens museum of Shaanxi Institute of Zoology, Xi’an, China. Genomic DNA was prepared in 150 bp paired-end libraries, tagged and subjected to the high-throughput Illumina Xten platform and yielded 19,970,960 Paired-End Raw Reads. Mapping against the complete mitogenome of M. moschiferus (GenBank: KT337321), high-quality reads were assembled using MITObim version 1.9 (Hahn et al. Citation2013). A total of 16,923 individual mitochondrial reads gave an average coverage of 154.7X. Comparing with the M. moschiferus, annotations were generated in MITOchondrial genome annotation Server (MITOS) (Bernt et al. Citation2013) and Geneious version 10.1.2.
The complete mitogenome sequence consists of 16,353 bp for M. berezovskii (GenBank: MH047347). The typical ATN (ATG or ATT or ATA) start codons are present in PCGs. TAA and AGA stop codons are used for most genes, with the exception of incomplete stop codon T for COX3, ND2 and ND4, and TA for ND3. The two rRNA genes are 955 bp in srRNA and 1571 bp in lrRNA. The tRNA genes have the typical cloverleaf secondary structures except for the shortest tRNASer(AGN) which lacks the DHU arm. A tandem repeat was not found in 924-bp-long non-coding region (Kim et al. Citation2017).
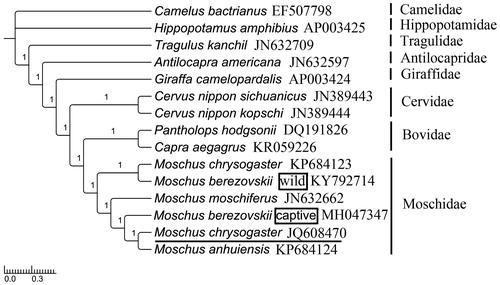
For phylogenetic analyses of Moschidae, MrBayes ver. 3.2.2 (Ronquist et al. Citation2012) and RAxML (Stamatakis Citation2006) were used to reconstruct BI and ML tree, understanding the best partitioned scheme and optimal model analysed in Partitionfinder v1.1.1 (Lanfear et al. Citation2012) (models GTR + I+G and GTR + G). Hippopotamus amphibius (GenBank: AP003425) and Camelus bactrianus (GenBank: EF507798) were selected as outgroups. The phylograms obtained from BI and ML (data not shown) all strongly indicated that Moschidae was a sister group to Bovidae (Hassanin and Douzery Citation2003; Yang et al. Citation2013; Pan et al. Citation2015). In Moschidae, our analysis supported that M. berezovskii should be divided into two subspecies, and the captive species was most closely related to the M. anhuiensis and the wild species was most closely related to M. chrysogaster (Su et al. Citation1999; ). Extraordinarily, the M. chrysogaster JQ608470 was located in the top of the tree, but not belonged to the branch of (M. chrysogaster KP684123, M. berezovskii wild) ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Gang Y, Wang BB, Ying Z, Wan QH, Fang SG. 2015. Low population density of the endangered forest musk deer, Moschus berezovskii, in China. Pak J Zool. 47:325–333.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Hassanin A, Douzery EJ. 2003. Molecular and morphological phylogenies of Ruminantia and the alternative position of the Moschidae. Syst Biol. 52:206–228.
- IUCN. 2017. The IUCN red list of threatened species. Version 2017-3; [accessed 2018 Mar 09]. www.iucnredlist.org.
- Kim SI, Lee MY, Jeon HS, Han SH, An J. 2017. Complete mitochondrial genome of Siberian musk deer Moschus moschiferus (Artiodactyla: Moschidae) and phylogenetic relationship with other Moschus species. Mitochondrial DNA B. 2:860–861.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Pan T, Wang H, Hu CC, Sun ZL, Zhu XX, Meng T, Meng XX, Zhang BW. 2015. Species delimitation in the genus Moschus (Ruminantia: Moschidae) and its high-plateau origin. PLoS One. 10:e0134183.
- Peng HY, Liu SC, Zou FD, Zeng B, Yue BS. 2008. Genetic diversity of captive forest musk deer (Moschus berezovskii) inferred from the mitochondrial DNA control region. Anim Genet. 40:65–72.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Su B, Wang YX, Lan H, Wang W, Zhang YP. 1999. Phylogenetic study of complete cytochrome b genes in musk deer (genus Moschus) using museum samples. Mol Phylogenet Evol. 12:241–249.
- Yang CZ, Xiang CK, Zhang XY, Yue BS. 2013. The complete mitochondrial genome of the Alpine musk deer (Moschus chrysogaster). Mitochondrial DNA A. 24:501–503.