Abstract
Notholirion macrophyllum (D. Don) Boiss. (Liliaceae) is a floriferous species naturally distributed in Asia. The complete chloroplast genome sequence of N. macrophyllum was generated by de novo assembly using whole genome next generation sequencing data. The complete chloroplast genome of N. macrophyllum was 152143 bp in total sequence length and divided into four distinct regions: small single copy region (17913 bp), large single copy region (82222 bp) and a pair of inverted repeat regions (26004 bp). The genome annotation displayed a total of 135 genes, including 82 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. Phylogenetic analysis with 7 Liliaceae species revealed that N. macrophyllum was the basal species of tribe Lilieae and was close to Cardiocrinum giganteum.
Keywords:
Notholirion macrophyllum (D. Don) Boiss. (Liliaceae) is a floriferous herb, which naturally distributes in high altitude region. It wildly grows in quercus forests, grassy slopes and meadows with an altitude in 2800–3400 m (Liang et al. Citation2000). Its flowers are gorgeous, and similar to lily. Notholirion macrophyllum also is used in traditional Chinese medicine (Liang et al. Citation2000). Notholirion Wall. ex Boiss. is a small Asian genus with only 5 species (Liang Citation1995). There were some researches hypothesized that Notholirion is the basal group of tribe Lilieae (Zhou Citation2008; Gao et al. Citation2012, Citation2013). As chloroplast carry maternal genes, it plays an important role in phylogeny reconstruction. However, there are no researches about Notholirion chloroplast. In this paper, we first report the complete chloroplast genome of N. macrophyllum, which will help in molecular and phylogenetical studies of this plant.
Fresh leaves of N. macrophyllum was collected from Daocheng (28°49′33″N 100°27′20″E), Sichuan Province, China. Voucher specimens were deposited in SZ (Sichuan University Herbarium). Total genomic DNA was extracted by Plant Genomic DNA Kit (Tiangen Biotech CO., LTD, Beijing, China). The isolated genomic was manufactured to average 350 bp paired-end(PE) library using Illumina HiSeq platform (Novogene, Beijing, China), and sequenced by Illumina genome analyser (Hiseq PE150). We found Cardiocrinum giganteum (Wall.) Makino was the best inference of nuclear genome to contribute for assembly. Contigs, assembled by SOAPdenovo2 (Luo et al. Citation2012), were sorted and joined into a single-draft sequence using Geneious (Kearse et al. Citation2012), which compared with the chloroplast sequence of C. giganteum as a reference. Gapcloser was used to fill the gapped sites, and the draft sequence was corrected manually by clean read mapping using bowtie2 (Langmead and Salzberg Citation2012) and Tablet (Milne et al. Citation2013). The genes in chloroplast genome were predicted using Geneious and corrected manually.
The complete chloroplast genome of N. macrophyllum (GenBank accession number MH011354) was 152,143 bp in total sequence length with 37.10% GC contents. Four distinct regions were separated by the complete chloroplast, such as large single copy (LSC) region was 82,222 bp, small single copy (SSC) region was 17,913 bp, and a pair of inverted repeat regions are 26,004 bp in each length. The chloroplast genome detected a total of 135 genes including 82 protein-coding genes, 38 tRNA genes, and 8 rRNA genes.
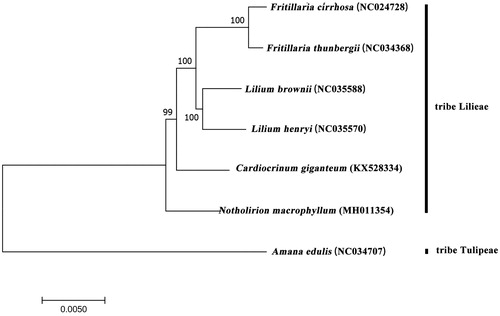
Aims at clarifying the phylogenetic relationship between N. macrophyllum with other Liliaceae species, we generated a maximum-likelihood tree (ML) of 7 species () by MEGA6 (Tamura et al. Citation2013). There are 6 species, which belongs to tribe Lilieae (Takhtajan Citation1987), Amana edulis (Miq.) Honda pertain to tribe Tulipeae. As shown in , N. macrophyllum was the basal group of the tribe Lilieae, and close to C. giganteum, these results in accordance with early studies (Zhou Citation2008; Gao et al. Citation2012, Citation2013).
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Gao YD, Harris AJ, Zhou SD, He XJ. 2013. Evolutionary events in Lilium (including Nomocharis, Liliaceae) are temporally correlated with orogenies of the Q-T plateau and the Hengduan Mountains. Mol Phylogenet Evol. 68:443–460.
- Gao YD, Zhou SD, He XJ. 2012. Chromosome diversity and evolution in tribe Lilieae (Liliaceae) with emphasis on Chinese species. J Plant Res. 125:55–69.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–U354.
- Liang S-Y. 1995. Chorology of Liliaceae(s.str.) and its bearing on the Chinese flora. Acta Phytotaxonomica Sinica. 33:27–51.
- Liang S, Minoru N. Tamura 2000. Notholirion Wallich ex Boissier. Flora of China. 24:133–134.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.
- Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 14:193–202.
- Takhtajan A. 1987. Systema Magnoliophytorum. Leinopli: Nauka
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhou SD. 2008. Phylogenetic and evolution of Lilieae (Liliaceae s.str.) in china. PhD thesis. Sichuan: University of Sichuan.