Abstract
Lilium pardalinum Kellogg is native to the Pacific Coast of the United States, and grows in woodland near streams. In the present study, the complete plastome of L. pardalinum was sequenced. The plastome sequence is 151,969 bp long with a large single copy, a small single copy, and two inverted repeat regions of length 81,401, 17,346, and 26,611 bp, respectively. A total of 133 genes were identified, including 82 coding genes, 8 ribosomal RNAs, 38 transfer RNAs, and 5 pseudogenes. Among the 5 pseudogenes, pseudo ndhF and ndhG genes were similar to that of L. washingtonianum and L. philadelphicum, respectively. Lilium pardalinum is sister to American lilies, except L. philadelphicum.
Keywords:
The section Pseudolirium Endlicher of the genus Lilium consists of ca. 20 species inhabiting North America. This section was considered to be very closely related to section Martagon because of whorled leaves (Comber Citation1949; Lighty Citation1968). However, the most recent phylogenetic analyses using molecular markers indicated that both the sections are distantly related (Hayashi and Kawano Citation2000; Lee et al. Citation2011; Gao et al. Citation2013; Du et al. Citation2014). Lilium pardalinum Kellogg belongs to the section Pseudolirium and is native to the Pacific Coast of the United States, growing in woodland near streams (McRae et al. Citation1998). In contrast to certain species in the same section, L. pardalinum has been successfully used for intersectional hybridization of the genus Lilium (van Tuyl and Arens Citation2010). However, sectional delimitation based on morphological and geographical characters of the genus Lilium has been controversial because the recent phylogenetic studies have not strongly supported the sectional classification of the genus Lilium (Hayashi and Kawano Citation2000; Du et al. Citation2014). Therefore, the genetic studies of Lilium species are necessary for systematics and prospective breeding of the genus Lilium. In the present study, the plastome of L. pardalinum was completely sequenced. The total genomic DNA was extracted from the fresh leaves of L. pardalinum using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). Seeds of L. pardalinum were collected at Onion Meadow, Tulare County, California on 28 September 1989 and grew in Kyungpook University, Korea. The living material and DNA were stored in Kyungpook University, Korea. The total DNA was sequenced using HiSeq 2500 instrument (Illumina, San Diego, CA, USA). All genes were annotated using Geneious (Kearse et al. Citation2012) by comparing with the previously reported plastome sequences of Lilium species. The phylogenetic analysis was performed with the concatenated 78 genes using randomized axelerated maximum likelihood (RAxML) (Stamatakis Citation2014). The plastome sequences of 22 Lilium and 3 Fritillaria species were downloaded from the National Center for Biotechnology Information database.
The plastome sequence is 151,969 bp long and consists of a large single copy region (81,401 bp), a small single copy region (17,346 bp), and two inverted repeat regions (26,611 bp). A total of 133 genes were identified, including 82 coding genes, 8 ribosomal RNAs, 38 transfer RNAs, and 5 pseudogenes.
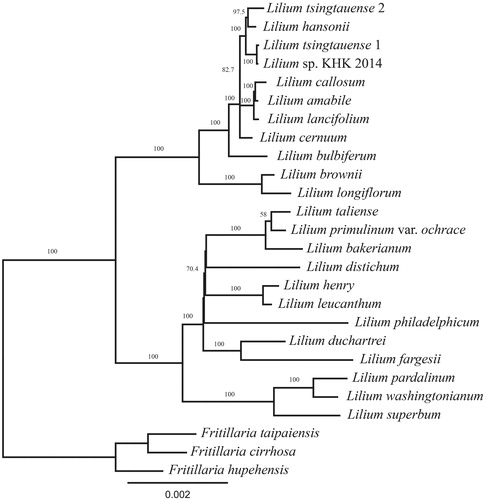
The phylogenetic analysis showed that L. pardalinum and L. washingtonianum formed a clade, and they were sister to L. superbum of the section Pseudolirium (). However, these three species were distantly related to L. philadelphicum. This polyphyly of the section Pseudolirium has been reported by previous studies (Hayashi and Kawano Citation2000; Lee et al. Citation2011); however, certain studies have supported that the section Pseudolirium forms a clade (Nishikawa et al. Citation2001; Du et al. Citation2014). However, our result does not imply that L. philadelphicum should be distinguished from other Pseudolirium species because there are still taxonomic gaps and questions on the misidentification of samples. Consequently, more phylogenetic studies will help us understand the relationship among the species of the genus Lilium.
Figure 1. Phylogenetic analysis with 78 genes using randomized axelerated maximum likelihood (RAxML). Lilium pardalinum and L. washingtonianum form a clade, and they are sister to L. superbum of the section Pseudolirium. However, L. philadelphicum, another species of the section Pseudolirium, is distant from rest of the section Pseudolirium. The numbers on the node and scale refer to bootstrap values and substitutions per site, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Comber HF. 1949. A new classification of the genus Lilium. Vol. 13. Lily Year Book, Royal Horticultural Society.13:86–105.
- Du YP, He HB, Wang ZX, Li S, Wei C, Yuan XN, Cui Q, Jia GX. 2014. Molecular phylogeny and genetic variation in the genus Lilium native to China based on the internal transcribed spacer sequences of nuclear ribosomal DNA. J Plant Res. 127:249–263.
- Gao YD, Harris AJ, Zhou SD, He XJ. 2013. Evolutionary events in Lilium (including Nomocharis, Liliaceae) are temporally correlated with orogenies of the Q-T plateau and the Hengduan Mountains. Mol Phylogenet Evol. 68:443–460.
- Hayashi K, Kawano S. 2000. Molecular systematics of Lilium and allied genera (Liliaceae): phylogenetic relationships among Lilium and related genera based on the rbcL and matK gene sequence data. Plant Species Biol. 15:73–93.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lee CS, Kim SC, Yeau SH, Lee NS. 2011. Major lineages of the genus Lilium (Liliaceae) based on nrDNA ITS sequences, with special emphasis on the Korean species. J Plant Biol. 54:159–171.
- Lighty R. 1968. Evolutionary trends in lilies. Lily Year Book RHS; 31:40–44.
- McRae EA, Austin-Mcrae E, MacRae E. 1998. Lilies: a guide for growers and collectors. Vol. 105. Portland: Timber Press.
- Nishikawa T, Okazaki K, Arakawa K, Nagamine T. 2001. Phylogenetic analysis of section Sinomartagon in genus Lilium using sequences of the internal transcribed spacer region in nuclear ribosomal DNA. Breed Sci. 51:39–46.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- van Tuyl JM, Arens P. 2011. Lilium: Breeding history of the modern cultiva assortment. Acta Hort. 900:223–230.
