Abstract
Fucus spiralis L. is a broadly distributed monoecious intertidal seaweed. The specific status of F. spiralis however is debatable. Here, we contribute to the bioinformatics and systematics of F. spiralis by analysing the complete mitochondrial and plastid genomes of a specimen from California, U.S.A. The F. spiralis mitogenome is 36,396 base pairs (bp) in length and contains 67 genes, and the plastid genome is 125,066 bp in length and contains 171 genes. The F. spiralis genomes are 99.7% and 99.8% similar in nucleotide sequence to F. vesiculosus, and support the revised classification of F. spiralis to Fucus vesiculosus var. spiralis.
Fucus spiralis is a common intertidal rockweed that occurs in the Atlantic and northeastern Pacific Oceans, and the Mediterranean Sea (Guiry and Guiry Citation2018). Hybridization studies demonstrate that F. spiralis forms reproductively successful hybrids with the closely related species F. vesiculosus, which exhibit no significant decrease in hybrid fertility (Kniep Citation1925; Burrows and Lodge Citation1951; Billard et al. Citation2005). Molecular phylogenetic analyses of the two species yield polytomies, and DNA sequences of accepted species markers find that F. spiralis differs from F. vesiculosus by as little as 0 bp for both cox1 and the internal transcribed spacer regions (Serrão et al. Citation1999; Coyer et al. Citation2006, Citation2011; Kucera and Saunders Citation2008; Laughinghouse et al. Citation2015). The only diagnostic feature appears to be highly polymorphic microsatellite markers (Engel et al. Citation2003; Wallace et al. Citation2004; Billard et al. Citation2005). In this study, we characterize the organellar genomes of F. spiralis to further understand its relationship to F. vesiculosus.
Fucus spiralis (Voucher Specimen – UC2050586) was collected from Pacific Grove, California (36°38′02.0″N, 121°56′19.7″W); its DNA was isolated following Lindstrom et al. (Citation2011). The 150 bp paired-end library construction and sequencing was performed by myGenomics, LLC (Alpharetta, GA). The genomes were assembled by mapping the reads against F. vesiculosus (GenBank – FM957154, AY494079) with the Low Sensitivity/Fast setting in Geneious R11 (Biomatters Limited, Auckland, New Zealand) and annotated using Sequin software. The mitogenome was aligned with other Phaeophyceae using MAFFT (Katoh and Standley Citation2013). The RaxML analysis was executed using complete mitogenome sequences at Trex-online (Boc and Makarenkov Citation2012) with the GTR + gamma model and 1000 fast bootstraps, then visualized with TreeDyn 198.3 at Phylogeny.fr (Dereeper et al. Citation2008).
The F. spiralis mitogenome (GenBank – MG922856) is 36,396 bp in length and contains 3 rRNA, 26 tRNA, and 28 other protein-coding genes. Its gene content and organization are the same as F. vesiculosus (Oudot-Le Secq et al. Citation2006). Fucus spiralis differs in sequence from F. vesiculosus by only 114 nucleotide SNPs and 10 gaps (99.7% similar). Comparison of all 9,367 protein coding amino acids finds 18 amino acid substitutions between the two species, of which only nine are radical (=the physiochemical properties are altered). Phylogenetic analysis of F. spiralis positions it in a fully supported clade with F. vesiculosus (). The plastid genome (GenBank – MG922855) is 125,066 bp in length and contains duplicate copies of 16S, 23S and 5S rRNAs, 26 tRNAs, and 139 protein-coding genes. It is also highly similar to F. vesiculosus in chromosomal content and structure (Le Corguillé et al. Citation2009). Fucus spiralis differs in sequence from F. vesiculosus by 234 nucleotide SNPs and 67 gaps (99.8% similar), and shows 168 amino acid substitutions out of 31,893 total amino acids, of which only 32 were radical.
Figure 1. Maximum-likelihood phylogram of Fucus vesiculosus var. spiralis (MG922856) and related Phaeophyceae mitogenomes. Numbers along branches are RaxML bootstrap supports based on 1000 nreps. The legend below represents the scale for nucleotide substitutions.
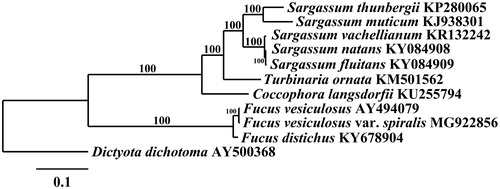
On the basis of this genomic data and evidence from the biological, marker, and phylogenetic species concepts, we conclude that the name F. spiralis should be reduced to varietal status under F. vesiculosus, F. vesiculosus var. spiralis (Linnaeus) Roth.
Acknowledgements
We wish to thank Long Nguyen for technical support with the Geneious software. This work was partly funded by a 2017–2018 Innovation Grant from Hartnell College to Jeffery R. Hughey.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Billard E, Daguin C, Pearson G, Serrão E, Engel C, Valero M. 2005. Genetic isolation between three closely related taxa: Fucus vesiculosus, F. spiralis, and F. ceranoides (Phaeophyceae). J Phycol. 41:900–905.
- Boc A, Makarenkov V. 2012. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40:W573–W579.
- Burrows EM, Lodge S. 1951. Autecology and the species problem in Fucus. J Mar Biol Assoc UK. 30:161–176.
- Coyer JA, Hoarau G, Oudot-Le Secq MP, Stam WT, Olsen JL. 2006. A mtDNA-based phylogeny of the brown algal genus Fucus (Heterokontophyta; Phaeophyta). Mol Phylogenet Evol. 39:209–222.
- Coyer JA, Hoarau G, Van Schaik J, Luijckx P, Olsen JL. 2011. Trans‐Pacific and trans‐Arctic pathways of the intertidal macroalga Fucus distichus L. reveal multiple glacial refugia and colonizations from the North Pacific to the North Atlantic. J Biogeogr. 38:756–771.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469.
- Engel CR, Brawley SH, Edwards KJ, Serrão EA. 2003. Isolation and cross-species amplification of microsatellite loci from the fucoid seaweeds Fucus vesiculosus, Fucus serratus and Ascophyllum nodosum (Heterokontophyta, Fucaceae). Mol Ecol Notes. 3:180–182.
- Guiry MD, Guiry GM. 2018. AlgaeBase [Internet]. Galway: World-Wide Electronic Publication, National University of Ireland; [cited 2018 Mar 26]. http://www.algaebase.org.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kniep H. 1925. Über Fucusbastarde. Flora. 118:331–338.
- Kucera H, Saunders GW. 2008. Assigning morphological variants of Fucus (Fucales, Phaeophyceae) in Canadian waters to recognized species using DNA barcoding. Botany. 86:1065–1079.
- Laughinghouse IVHD, Müller KM, Adey WH, Lara Y, Young R, Johnson G. 2015. Evolution of the northern rockweed, Fucus distichus, in a regime of glacial cycling: implications for benthic algal phylogenetics. PLoS One. 10:e0143795.
- Le Corguillé G, Pearson G, Valente M, Viegas C, Gschloessl B, Corre E, Bailly X, Peters AF, Jubin C, Vacherie B, et al. 2009. Plastid genomes of two brown algae, Ectocarpus siliculosus and Fucus vesiculosus: further insights on the evolution of red-algal derived plastids. BMC Evol Biol. 9:253.
- Lindstrom SC, Hughey JR, Martone PT. 2011. New, resurrected and redefined species of Mastocarpus (Phyllophoraceae, Rhodophyta) from the northeast Pacific. Phycologia. 50:661–683.
- Oudot-Le Secq MP, Loiseaux-de Goer S, Stam WT, Olsen JL. 2006. Complete mitochondrial genomes of the three brown algae (Heterokonta: Phaeophyceae) Dictyota dichotoma, Fucus vesiculosus and Desmarestia viridis. Curr Genet. 49:47–58.
- Serrão EA, Alice LA, Brawley SH. 1999. Evolution of the Fucaceae (Phaeophyceae) inferred from nrDNA-ITS. J Phycol. 35:382–394.
- Wallace AR, Klein AS, Mathieson AC. 2004. Determining the affinities of salt marsh fucoids using microsatellite markers: evidence of hybridization and introgression between two species of Fucus (Phaeophyceae) in a Maine estuary. J Phycol. 40:1013–1027.
