Abstract
The Clausena lansium, originally native to the southern part of China and has a long history of cultivation, is a tree member of the family Rutaceae. Chloroplast genome sequences play a significant role in the development of molecular markers in plant phylogenetic and population genetic studies. In this study, we report the complete chloroplast genome sequence of C. lansium for the first time (accession number of MH047377). The chloroplast genome is 159,284 bp long and includes 113 genes. It’s LSC, SSC, and IR regions are 88,634, 18,896, and 25,877 bp long, respectively. Phylogenetic tree analysis exhibited that C. lansium was clustered with other Rutaceae species with high bootstrap values.
Wampee (Clausena Burm. F.) is a tropical and subtropical, very remote citroid fruit tree belonging to subtribe Clauseninae, tribe Clauceneae of the family Rutaceae (Swingle and Reece Citation1967). Wampee has more than 30 species, 11 of which are native to China (Yu Citation1982). Among them, only Clausena lansium (Lour.) Skeels and C. indica (Dalz.) Oliv. are edible and commercially cultivated. Chinese wampee (C. lansium) was originally native to the southern part of China and has a long history of cultivation. Presently, its large commercial cultivation areas are the Guangdong, Guangxi and Fujian Provinces, and ‘Chicken Heart’ sweet wampee is the most famous cultivar. Wampee is a remote relative of Citrus, and sexual incompatibility exists between them (Guo and Deng Citation1999), but the phylogenetic relationship between them is unclear.
In this study, we report the complete chloroplast genome of C. lansium, the second complete plastome sequence from the genus Clausena. DNA material was isolated from mature leaves of a C. lansium plant cultivated in the plant garden of Yunnan Institute of Tropical Crops (YITC), Jinghong, China by using DNeasy Plant Mini Kit (QIAGEN, Germany). A specimen of this tree and the isolated DNA were stored in YITC. About 10 μg isolated DNA was sent to BGI, Shenzhen for library construction and genome sequencing on the Illumina Hiseq 2000 Platform. After genome sequencing, a total of 4.5 Gbp reads in fastq format were obtained and subjected to chloroplast genome assembly. The complete chloroplast genome was annotated with Dual Organelle GenoMe Annotator (DOGMA; Wyman et al. Citation2004) and submitted to the Genbank under the accession number of MH047377. Our assembly of the C. lansium resulted in a final sequence of 159,284 bp in length with no gap. The overall A–T content of the chloroplast genome was 61.1%. This chloroplast genome included a typical quadripartite structure with the Large Single Copy (LSC), Small Single Copy (SSC), and Inverted Repeat (IR) regions of 88,634, 18,896, and 25,877 bp long, respectively. Genome annotation showed 113 full length genes including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The genome organization, gene content, and gene relative positions were almost identical to the previously reported Rutaceae chloroplast genomes (Liu and Shi Citation2017).
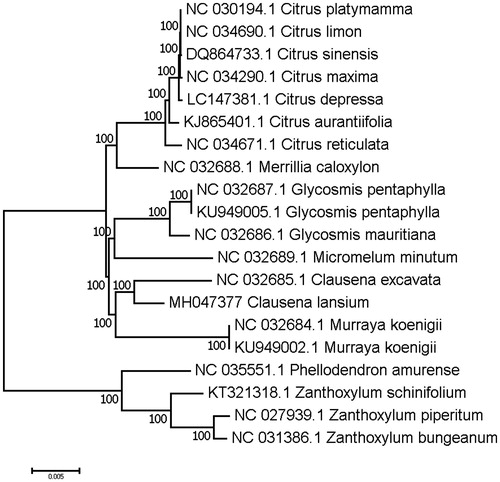
To validate the phylogenetic relationships of C. lansium in the Rutaceae, we constructed a maximum likelihood tree using 20 Rutaceae taxa. Phylogenetic analysis was performed on a data set that included 79 protein-coding genes and 4 rRNA genes from the 20 selected taxa using RAxML v. 7.7.1 (Stamatakis et al. Citation2008). The 83 gene sequences (82,566 bp in length) were aligned with the MAFFT (Katoh and Standley Citation2013). The resulting tree shows that C. lansium forms a clade with the species of C. excavata, and sister to Murraya koenigii with a 100% bootstrap value ().
Disclosure statement
The authors declare that they have no conflict of interest.
Additional information
Funding
References
- Guo WW, Deng XX. 1999. Intertribal hexaploid somatic hybrid plants regeneration from electrofusion between diploids of citrus sinensis and its sexually incompatible relative, Clausena lansium. Theor Appl Genet. 98:581–585.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Liu J, Shi C. 2017. The complete chloroplast genome of wild shaddock, citrus maxima, (burm.) merr. Conserv Genet Resour. 9:1–3.
- Swingle WT, Reece PC. 1967. The botany of citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD, editors. The citrus industry. Vol. 1. Berkeley: University of California Press; p. 190–430.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Yu DJ. 1982. Taxonomy of fruit crops in China (in Chinese). Beijing: Press of Agriculture; p. 338–344.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20:3252–3255.