Abstract
The mitochondrial genome of Fejervarya kawamurai is a circular molecule of 17,650 bp in length, containing 13 protein-coding genes, two rRNA genes, 23 tRNA genes (including an extra tRNA-Met), and the control region. The AT content of the whole genome is 56.9%. In Bayesian inference (BI) and Maximum likelihood (ML) analyses, we found that F. kawamurai is a sister clade to F. multistriata and F. limnocharis. The monophyly of Fejervarya, Quasipaa, Nanorana was well supported (1.00 in BI and 100% in ML).
Fejervarya kawamurai was described as a new species of dicroglossid frog and a member of the Fejervarya limnocharis complex from western Honshu, Japan mainland by Djong et al. (Citation2011). It is distributed broadly across parts of Japan, Taiwan, and China (Frost Citation2018). There is much controversy about the classification of F. kawamurai and the closely related F. limnocharis and F. multistriata (Djong et al. Citation2011; Huang and Tu Citation2016). The complete mitochondrial genomes of F. limnocharis and F. multistriata were sequenced but the comparable genome of F. kawamurai was unknown. Hence, we sequenced the mitochondrial genome of F. kawamurai to discuss the relationship within the Fejervarya limnocharis complex.
Samples of F. kawamurai were collected from Zunyi city, Guizhou province, China (27°42′30′′, 106°55′12′). Whole genomic DNA was extracted from the hind leg muscle. The frog’s samples and DNA samples were stored at the College of Chemistry and Life Science, Zhejiang Normal University, China. DNA fragments were amplified using 15 pairs of highly conserved primers for mitochondrial genes which were designed according to the method of Liu et al. (Citation2005) and Huang and Tu (Citation2016). All PCR procedures were performed using an Arktik Thermal Cycler (Thermal Scientific,Shanghai, China). The PCR products were sequenced by Sangon Biotech Company (Shanghai, China).
The complete mitochondrial genome of F. kawamurai is 17,650 bp in length, containing 13 protein-coding genes, two rRNA genes, 23 tRNA genes (an extra tRNA-Met is present), and the control region. The AT content of the complete mtDNA is 56.9%. Most protein-coding genes begin with ATG as the start codon, except for ND5 gene with GTC, ND2 gene with ATT, COI gene with ATA, and ND3 gene with GTG. The ND6 gene is terminated with AGG as the stop codon, whereas ND1, ND2, ND5, COI, COII, ATP6, and COIII genes end with an incomplete stop codon (T–) and the other protein-coding genes end with TAA.
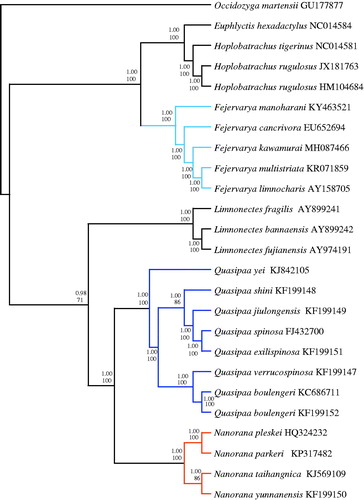
Bayesian inference (BI) and maximum likelihood (ML) trees were constructed using the 13 protein-coding genes of 25 species (Liu et al. Citation2005; Ren et al. Citation2009; Zhang et al. Citation2009; Zhou et al. Citation2009; Alam et al. Citation2010; Chen et al. Citation2011; Yu et al. Citation2012; Shan et al. Citation2014; Yu et al. Citation2015; Chen et al. Citation2015a,Citationb; Jiang et al. Citation2016; Kiran et al. Citation2017), including Occidozyga martensii (Li et al. Citation2014) as outgroup to confirm the phylogenetic position of F. kawamurai in Dicroglossidae (). To select conserved regions of the putative nucleotide sequences, each alignment was analyzed with the program Gblocks 0.91 b (Castresana Citation2000) using default settings. BI analysis and ML analysis was performed by MrBayes 3.1.2 (Huelsenbeck and Ronquist Citation2001) and RaxML HPC (Stamatakis et al. Citation2008), respectively. In both the BI and ML trees, we found that F. kawamurai is a sister clade to (F. multistriata + F. limnocharis). The monophyly of Fejervarya, Quasipaa, Nanorana was well supported (1.00 in BI and 100% in ML) as also reported in other recent studies (Zhang et al. 2009; Cai et al. Citation2018).
Figure 1. Phylogenetic relationships among 25 species of Dicroglossidae based on 13 protein-coding genes using nucleotide datasets. The tree was rooted with Occidozyga martensii as the out-group. Numbers above the nodes are the posterior probabilities of BI on top and the bootstrap values of ML on the bottom.
Nucleotide sequence accession number
The complete mitochondrial genome of F. kawamurai has been assigned the GenBank accession number MH087466.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Alam MS, Kurabayashi A, Hayashi Y, Sano N, Khan MMR, Fujii T, Sumida M. 2010. Complete mitochondrial genomes and novel gene rearrangements in two dicroglossid frogs, Hoplobatrachus tigerinus and Euphlyctis hexadactylus, from Bangladesh. Genes Genet Syst. 85:219–232.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552.
- Cai YT, Ma L, Xu CJ, Li P, Zhang JY, Storey KB, Yu DN. 2018. The complete mitochondrial genome of the hybrid of Hoplobatrachus chinensis (♀) × H. rugulosus (♂) and its phylogeny. Mitochondrial DNA B. 3:344–345.
- Chen G, Wang B, Liu J, Xie F, Jiang JP. 2011. Complete mitochondrial genome of Nanorana pleskei (Amphibia: Anura: Dicroglossidae) and evolutionary characteristics of the amphibian mitochondrial genomes. Cur Zool. 57:785–805.
- Chen Z, Zhai X, Zhang J, Chen X. 2015a. The complete mitochondrial genome of Feirana taihangnica (Anura: Dicroglossidae). Mitochondrial DNA. 26:485–486.
- Chen Z, Zhai X, Zhu Y, Chen X. 2015b. Complete mitochondrial genome of the Ye’s spiny-vented frog Yerana yei (Anura: Dicroglossidae). Mitochondrial DNA. 26:489–490.
- Djong HT, Matsui M, Kuramoto M, Nishioka M, Sumida M. 2011. A new species of the Fejervarya limnocharis complex from Japan (Anura, Dicroglossidae). Zool Sci. 28:922–929.
- Frost DR. 2018. Amphibian Species of the World: an Online Reference. American Museum of Natural History, New York, USA. Accessible at http://research.amnh.org/herpetology/amphibia/index.html [accessed 1 March 2018].
- Huang ZH, Tu FY. 2016. Mitogenome of Fejervarya multistriata: a novel gene arrangement and its evolutionary implications. Genet Mol Res. 15:1–9.
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Jiang L, Ruan Q, Chen W. 2016. The complete mitochondrial genome sequence of the Xizang Plateau frog, Nanorana parkeri (Anura: Dicroglossidae). Mitochondrial DNA Part A. 27:3184–3185.
- Kiran SK, Anoop VS, Sivakumar KC, Dinesh R, Mano JP, Kaushik D, Sanil G. 2017. An additional record of Fejervarya manoharani Garg and Biju from the Western Ghats with a description of its complete mitochondrial genome. Zootaxa. 4277:491.
- Li E, Li X, Wu X, Feng G, Zhang M, Shi H, Wang L, Jiang J. 2014. Complete nucleotide sequence and gene rearrangement of the mitochondrial genome of Occidozyga martensii. J Genet. 93:631–641.
- Liu ZQ, Wang YQ, Su B. 2005. The mitochondrial genome organization of the rice frog, Fejervarya limnocharis (Amphibia: Anura): a new gene order in the vertebrate mtDNA. Gene. 346:145–151.
- Ren Z, Zhu B, Ma E, Wen J, Tu T, Cao Y, Hasegawa M, Zhong Y. 2009. Complete nucleotide sequence and gene arrangement of the mitochondrial genome of the crab-eating frog Fejervarya cancrivora and evolutionary implications. Gene. 441:148–155.
- Shan X, Xia Y, Zheng YC, Zou FD, Zeng XM. 2014. The complete mitochondrial genome of Quasipaa boulengeri (Anura: Dicroglossidae). Mitochondrial DNA. 25:83–84.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Yu DN, Zhang JY, Li P, Zheng RQ, Shao C. 2015. Do cryptic species exist in Hoplobatrachus rugulosus? An examination using four nuclear genes, the Cyt b gene and the complete MT genome. PLoS One. 10:e0124825.
- Yu DN, Zhang JY, Zheng RQ, Shao C. 2012. The complete mitochondrial genome of Hoplobatrachus rugulosus (Anura: Dicroglossidae). Mitochondrial DNA. 23:336–337.
- Zhang JF, Nie LW, Wang Y, Hu LL. 2009. The complete mitochondrial genome of the large-headed frog, Limnonectes bannaensis (Amphibia: Anura), and a novel gene organization in the vertebrate mtDNA. Gene. 442:119–127.
- Zhou Y, Zhang JY, Zheng RQ, Yu BG, Yang G. 2009. Complete nucleotide sequence and gene organization of the mitochondrial genome of Paa spinosa (Anura: Ranoidae). Gene. 447:86–96.
