Abstract
Glycine soja (wild soybean) is known as the wild progenitor of cultivated soybean (G. max), representing a valuable genetic resource for soybean breeding programmers. In this study, we determined the complete mitochondrial genome of G. soja. The results revealed that G. soja mt genome is 402,545 bp with 45% GC contents. Furthermore, based on entire mitochondrial genome the evolutionary relationship and phylogenetic analysis revealed that G. soja closely related to G. max.
The mitochondrion is a membrane-enclosed organelle discovered in eukaryotic cells (Henze and Martin Citation2003). Plant mitochondrial DNA (mtDNAs) are a major resource for evolutionary studies, because coding regions evolve slowly, in contrast to the flexible non-coding DNA. Therefore, the structural evolution and plasticity of plant mtDNAs make them powerful model for exploring the forces that affect their divergence and recombination. Soybean (Glycine) is standout amongst the essential worldwide crops, grown for vegetable oil and protein. Soybean nuclear and chloroplast genomes had been published (Saski et al. Citation2005; Schmutz et al. Citation2010; Asaf et al. Citation2017), significantly increasing our expertise of soybean biology. The arrival of excessive-throughput sequencing technology has facilitated rapid developments in the field of genomics, especially in mitogenome genetics. In the present work, the complete mitochondrial genome of wild soybean G. soja was sequenced (GenBank accession number: MF955859) with the aim to know its phylogenetic position on the basis of entire mitogenomes.
The G. soja (accession KLG90379), seeds were received from the National GeneBank of the Rural Development Administration of the Republic of Korea. Mitochondrial DNA was extracted from young leaves using the method described previously (Asaf et al. Citation2016; Wu Citation2016). Approximately eighty million raw Illumina reads were demultiplexed and trimmed. The raw reads were filtered and then assembled de novo into contigs using Geneious Pro v11.1 (http://geneious.com). BLAST searches were conducted on all of the contigs using the NCBI database (http://www.ncbi.nlm.nih.gov/) for the annotation of mitochondrial sequences. To identify tRNAs IN the genome tRNA scan-SE software (http://lowelab.ucsc.edu/tRNAscan-SE/) was used. The complete mitochondrial genome was used to infer its phylogenetic position using Neighbour-Joining (Saitou and Nei Citation1987) tree with 1000 bootstrap replications (Felsenstein Citation1985).
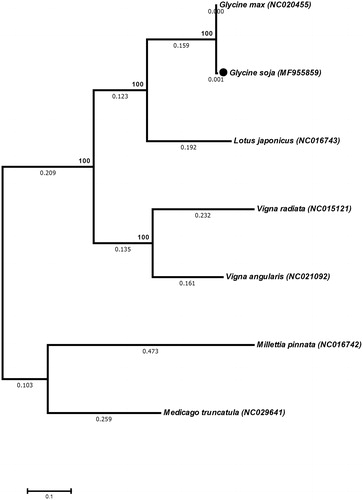
The complete mt genome of G. soja was 402,545 bp in size, encoded 111 genes, including 19 transfer RNA (trn), 3 ribosomal RNA (rrn) genes and 36 protein coding genes (eight subunits of NADH dehydrogenase, 3 subunits of cytochrome c oxidase and seven subunits of ATP synthase) (). Glycine soja mt genome is 13 bp less than previously reported G. max mt genome. Among these genes, atp1 and atp 6 genes are present in three and two copies, respectively. Furthermore, Like G. max mitochondrial genome G. soja also has 52 ORFs in it mitochondrial genome (Chang et al. Citation2013). Similarly, the phylogenetic analysis revealed that mt genome of G. soja is closely related to G. max (). This study will help to understand the evolution of G. soja mitochondrial genome within the Family Fabaceae.
Acknowledgments
This work was supported by Korea Institute of Planning and Evaluation for Technology in food, Agriculture, Forestry and Fisheries (IPET) through Agriculture, Food and Rural Affairs Research Center Support Program, Funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA)(716001-7).
Disclosure statement
The authors declare no competing financial interest. The authors alone are responsible for the content and writing of the paper.
References
- Asaf S, Khan AL, Khan MA, Imran QM, Kang S-M, Al-Hosni K, Jeong EJ, Lee KE, Lee I-J. 2017. Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PloS One. 12:e0182281.
- Asaf S, Khan AL, Khan AR, Waqas M, Kang S-M, Khan MA, Shahzad R, Seo C-W, Shin J-H, Lee I-J. 2016. Mitochondrial genome analysis of wild rice (Oryza minuta) and its comparison with other related species. PloS One. 11:e0152937.
- Chang S, Wang Y, Lu J, Gai J, Li J, Chu P, Guan R, Zhao T. 2013. The mitochondrial genome of soybean reveals complex genome structures and gene evolution at intercellular and phylogenetic levels. PLoS One. 8:e56502.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39:783–791.
- Henze K, Martin W. 2003. Evolutionary biology: essence of mitochondria. Nature. 426:127.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Saski C, Lee S-B, Daniell H, Wood TC, Tomkins J, Kim H-G, Jansen RK. 2005. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol. 59:309–322.
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J. 2010. Genome sequence of the palaeopolyploid soybean. Nature. 463:178.
- Wu Z. 2016. The whole chloroplast genome of shrub willows (Salix suchowensis). Mitochondrial DNA Part A. 27:2153–2154.