Abstract
The Pink dentex (Dentex gibbosus, Rafinesque 1810) is one of the most commercially important Sparidae species and it is often subjected to fraud. Here, we report the complete mitochondrial genome of D. gibbosus. The mitogenome is 16,771 bp in length and contained 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes and 2 non-coding regions. The overall base composition of D. gibbosus mtDNA is: 27.8% for A, 28.60% for C, 16.5% for G, 27.05% for T.
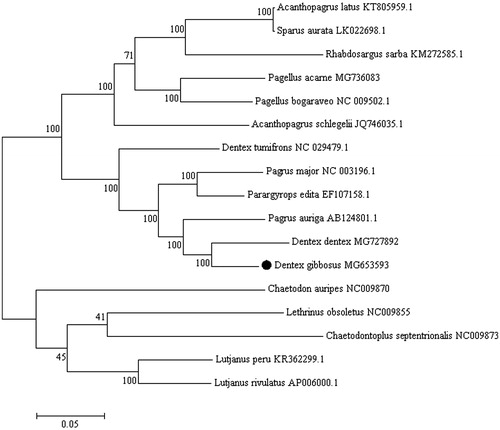
The Pink dentex (Dentex gibbosus) is a commercially important Sparidae species inhabiting the West African coast from Portugal to Angola, including Madeira, Canary and Sao Tome-Principe archipelagos (Wirtz et al. Citation2008). This sparid species is also present in the Mediterranean Sea except for the north-western coast and the northern Adriatic Sea (Dooley et al. Citation1985). Dentex gibbosus is now classified as “Least Concern” in the Red List of Threatened Species in the Mediterranean Sea (Russell et al. Citation2014). Species substitution is very common in processed fishery products belonging to Sparidae family. In particular, Dentex dentex and Pagrus pagrus, are often fraudulently replaced with D. gibbosus (Katavic et al. Citation2000). Here, we describe the complete mitochondrial genome (mitogenome) of D. gibbosus (GenBank MG653593). A specimen caught in the Mediterranean Sea (N 38°25′15.1″ E 3°53′09.4″) was identified as D. gibbosus based on morphological features. DNA was extracted and is currently stored at Department of Veterinary Medicine and Animal Production, University “Federico II”, Naples, Italy. The mitogenome of D. gibbosus has been obtained from high-throughput sequencing on complete mitochondrial DNA with Illumina HiSeq 2500 System (Illumina, San Diego, CA). The complete sequence is 16,771 bp long, containing 13 protein-coding genes, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA) and two non-coding regions (D-loop and L-origin). Mitochondrial arrangement and gene distribution are in agreement with the classic vertebrate mitogenomes (Wang et al. Citation2008). The majority of mitochondrial genes are encoded on the heavy strand, while the NADH dehydrogenase subunit 6 (ND6) and eight tRNA genes [Gln, Ala, Asn, Cys, Tyr, Ser(UCN), Glu, Pro] are encoded on the light strand. Base composition is similar to other Sparidae mitochondrial genomes, with 27.8% for A, 28.60% for C, 16.5% for G, and 27.05% for T (Ceruso et al. Citation2018). All the protein-coding genes started with an ATG start codon but COI and ND4, which started with GTG. Stop codons were of four types, i.e. TAA (ND1, ATP8, ATP6, ND4L, ND5, ND6), AGG (COI), T (COII, ND3, ND4, CYTB) and TA (ND2, COIII). The 12S and 16S rRNA genes were located between the tRNAPhe (GAA) and tRNALeu (TAA) genes, and were separated by the tRNAVal gene as in other vertebrates (Li et al. Citation2016). The 22 tRNA genes vary from 66 to 74 bp in length. The 1091 bp long control region is located between tRNAPro (TGG) and tRNAPhe (GAA). The non-coding region (L-strand origin of replication) is 40 bp long and is located between tRNAAsn (GTT) and tRNACys (GCA). To validate the phylogenetic position of D. gibbosus, we construct a phylogenetic tree using MEGA6 software (Tamura et al. Citation2013) (). The resultant phylogeny shows that D. gibbosus is closely related to D. dentex, in agreement with Chiba et al. (Citation2009). Results of this study provided useful genetic information for further studies on phylogeny, species identification and population genetics in Sparidae species.
Figure 1. Phylogenetic analysis of D. gibbosus based on the entire mtDNA genome sequences of 11 sparid fishes available in GenBank. Five outgroup species (Lutjanus peru, Lutjanus rivulatus, Lethrinus obsoletus, Chaetodontoplus septentrionalis and Chaetodon auripes) were selected and the maximum likelihood method was used. Numbers above the nodes indicate 1000 bootstrap values. Accession numbers are shown behind species names.
Acknowledgements
The authors thank the Molecular Biology and Sequencing unit of the Stazione Zoologica Anton Dohrn for technical assistance.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Funding
This work was supported by the Italian MIUR (Ministry of Education, University and Research) (PONa3_239), and Stazione Zoologica Anton Dohrn/University “Federico II” PhD fellowship to C.M.
References
- Ceruso M, Mascolo C, Palma G, Anastasio A, Pepe T, Sordino P. 2018. The complete mitochondrial genome of the common dentex, Dentex dentex (Perciformes: Sparidae). Mitochondrial DNA Part B. 3:391–392.
- Chiba SN, Iwatsuki Y, Yoshino T, Hanzawa N. 2009. Comprehensive phylogeny of the family Sparidae (Perciformes: Teleostei) inferred from mitochondrial gene analyses. Genes Genet Syst. 84:153–170.
- Dooley JK, Van Tassell JL, Brito A. 1985. An annotated checklist of the shorefishes of the Canary Islands. Am Museum Novitates. 2824:1–49.
- Katavic I, Grubisic L, Skakelja N. 2000. Growth performance of pink dentex as compared to four other sparids reared in marine cages in Croatia. Short Communication. Aquaculture International. 8:455–461.
- Li J, Yang H, Xie Z, Yang X, Xiao L, Wang X, Li S, Chen M, Zhao H, Zhang Y. 2016. The complete mitochondrial genome of the Rhabdosargus sarba (Perciformes: Sparidae). Mitochondrial DNA Part A. 27:1606–1607.
- Russell B, Carpenter KE, Pollard D. 2014. Dentex gibbosus. [accessed 2018 Jan 30]. The IUCN Red List of Threatened Species 2014:e.T170186A1289197. http://dx.doi.org/10.2305/IUCN.UK.20143.RLTS.T170186A1289197.en.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang C, Chen Q, Lu G, Xu J, Yang Q, Li S. 2008. Complete mitochondrial genome of the grass carp (Ctenopharyngodon idella, Teleostei): insight into its phylogenic position within Cyprinidae. Gene. 424:96–101.
- Wirtz P, Fricke R, Biscoito MJ. 2008. The coastal fishes of Madeira Island-new records and an annotated check-list. Zootaxa. 1715:1–26.
